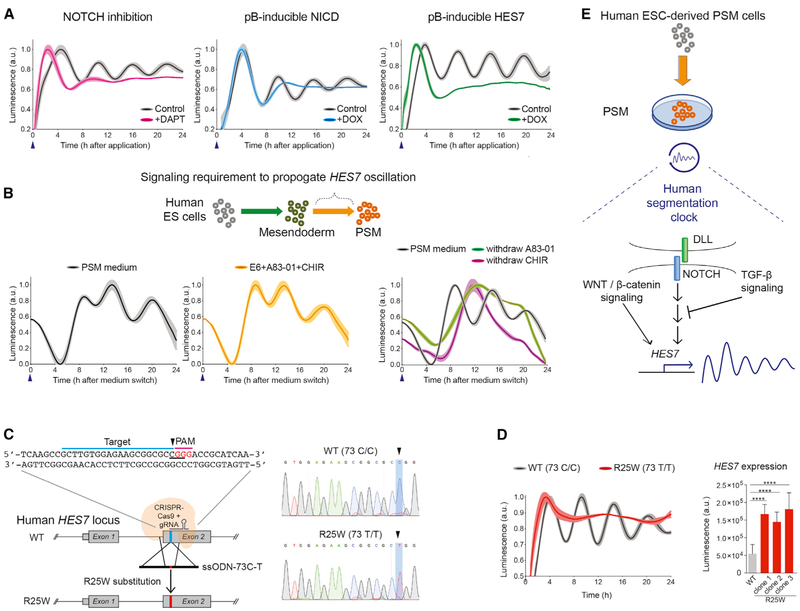
Figure 3. Characterization of HES7 Oscillation and Modeling SCDO4 with Genome Editing.
(A) The impact of modulating NOTCH signaling pathways on HES7 oscillation. Left panel: DAPT treatment (magenta). Middle panel: inducible NICD transgene expression (blue). Right panel: inducible HES7 transgene expression (green). The controls are shown in black. Blue arrowhead indicates when the NLuc luciferin and small molecules or DOX were applied. All luminescence oscillation data presented is scaled from minimum to maximum signal and shown as mean ± SEM (shaded area), obtained from 4 to 5 replicates in each experiment.
(B) A schematic for measuring the propagation of HES7 oscillation. The lower panels show the comparison between PSM medium (black) and with base medium supplemented with A83-01 and CHIR (yellow) only. Right panel: PSM medium (black) compared with only withdrawal of A83-01 (green) or the withdrawal of CHIR (magenta).
(C) Strategy for genome editing to introduce a R25W substitution. The sequences for gRNA protospacer adjacent motif (PAM) domain (magenta) and targeted sequences (blue) are indicated. The codon to be mutated is underlined, CGG (Arg) to TGG (Trp). Right panel: Sanger sequencing results of the parental line (wild type [WT]) and a homozygous R25W clone.
(D) Representative oscillation profile of WT (black) overlay with a R25W clone (red). Right panel: quantification of HES7 expression level between WT and R25W clones. ****p < 0.0001, Student’s t test. Data presented as the mean ± SD.
(E) A proposed working model of human segmentation clock and HES7 oscillation. NOTCH signaling is upstream of HES7 activation and oscillation. During the transition from mesendoderm to PSM state, WNT/β-catenin signaling activation and TGF-β signaling inhibition are crucial to propagate HES7 oscillation. Luminescence is measured in arbitrary units (a.u.).