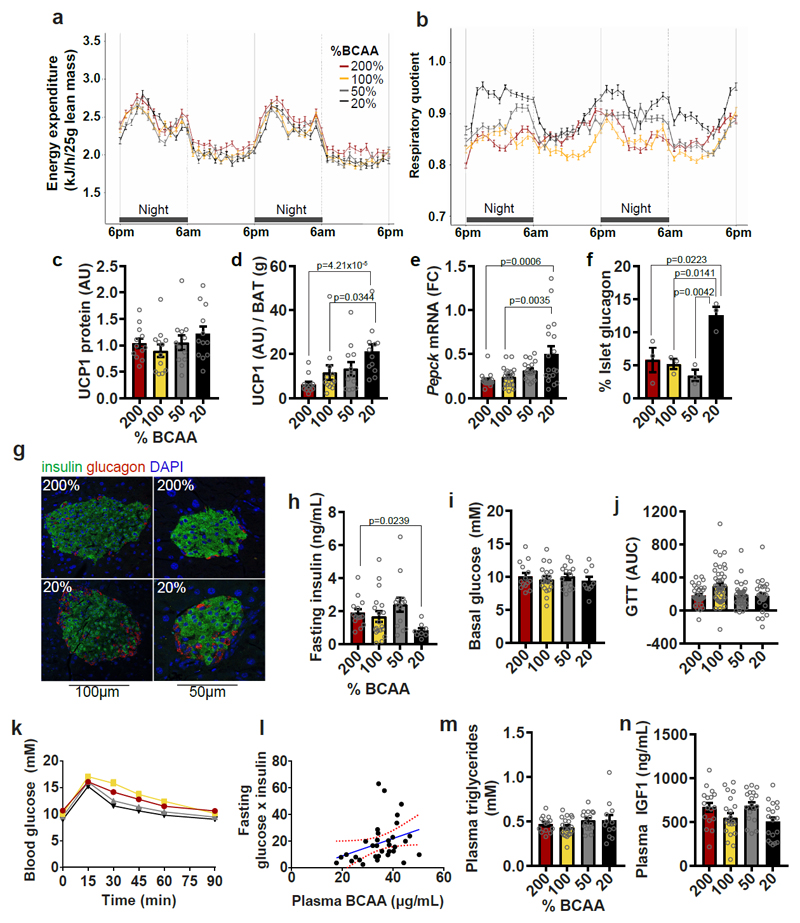
Fig. 6. Dietary amino acid imbalance alters whole body metabolism.
(a) Energy expenditure and (b) Respiratory quotient (RQ) (200%, 50% and 20%, n=12; 100%, n=16 biologically independent mice). Energy expenditure and RQ were measured in individual animals over 2 day and 2 night cycles using metabolic cages. (c) UCP1 protein expression in BAT (n=12 biologically independent mice) and (d) UCP1 protein per g of BAT mass (n=12 biologically independent mice). All western blots were standardized to contain 25ug of protein. (e) Pepck mRNA expression in liver (200%, n=17; 100%, n=21; 50% and 20%, n=18 biologically independent mice). (f) Glucagon content in pancreatic islets (n=3 mice) and (g) representative images of BCAA200 and BCAA20 islets visualized by immunohistochemistry, conducted once with 3 independent mice). Glucose metabolism is shown by (h) fasting insulin levels (200%, n=14; 100%, n=19; 50%, n=14; 20%, n=8 mice), (i) basal glucose levels (200%, n=14;, 100%, n=19; 50%, n=15; 20%, n=9 mice) and (j,k) the area under the curve (AUC) from glucose tolerance tests (GTT) (200%, n=33; 100%, n=44; 50%, n=33%; 20%, n=27 mice). (l) The relationship between plasma BCAAs and the product of fasting glucose and insulin, an index of insulin sensitivity (p=0.054; r=0.115; Pearson’s correlation; n=33 mice). Red lines show the 95% confidence interval. Plasma (m) triglycerides (200%, n=14, 100%, n=20; 50%, n=16; 20%, n=12 mice) and (n) IGF1 (200%, n=17; 100%, n=21, 50% and 20%, n=18 mice) were measured. For all bar graphs, ANOVA for normal and log-normal data, and Kruskal Wallis tests for non-normal data, were used to determine significant differences between groups. Pairwise comparisons amongst diets for normal and log-normal data were made using t-tests (two-sided). For non-normal data, pairwise comparisons amongst diets were made using Kruskal Wallis test. All bars indicate means ± SEM. *p≤0.05, **p≤0.01, ***p≤0.001 based on posthoc analysis following correction for multiple testing.