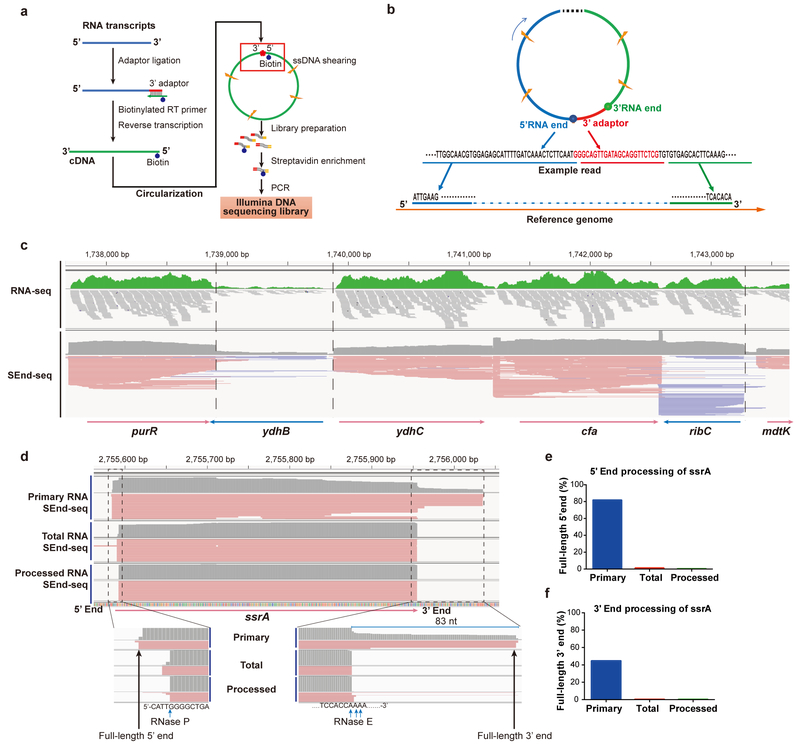
Fig. 1 |. Simultaneous capture of 5’- and 3’-end sequences of bacterial transcripts by SEnd-seq.
a, Workflow of SEnd-seq. See Methods for details. b, An example read illustrating how to infer the full-length sequence of individual transcripts by extracting correlated 5’- and 3’-end sequences and mapping them to the reference genome. c, A sample data track of the log-phase E. coli transcriptome showing the comparison between standard RNA-seq and SEnd-seq. Dashed lines highlight the sharp boundaries of transcripts delineated by SEnd-seq, which are obscured in standard RNA-seq. d, SEnd-seq reads mapped to the ssrA gene in primary, total and processed RNA datasets. e, Ratio of ssrA transcripts with an intact, unprocessed 5’ end in different datasets. f, Ratio of ssrA transcripts with an intact 3’ end in different datasets.