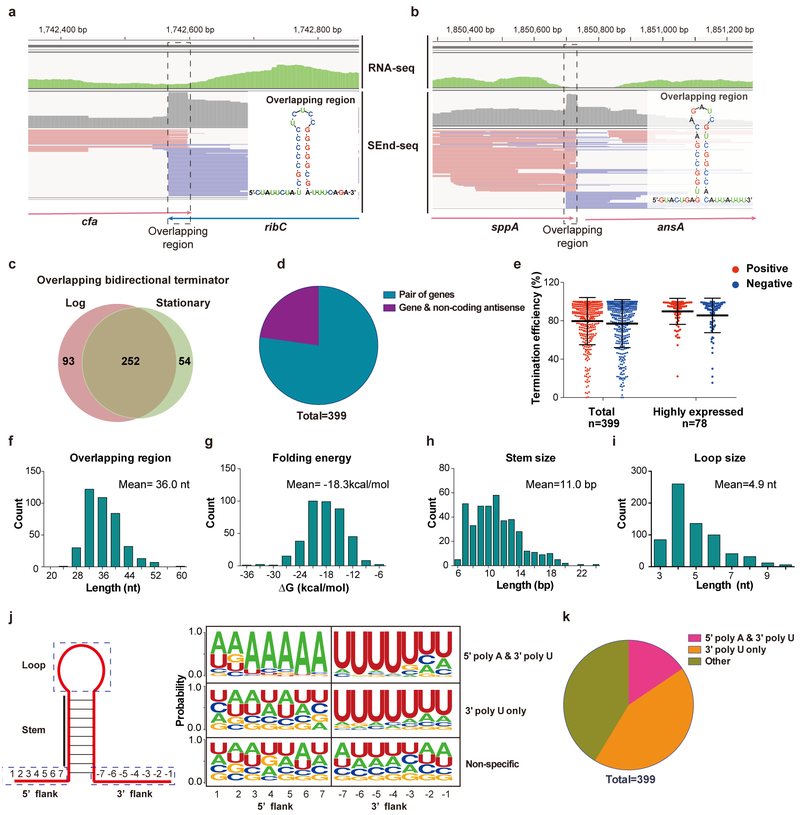
Fig. 4 |. Pervasive bidirectional overlapping TTS revealed by SEnd-seq.
a, SEnd-seq data track for an example convergent gene pair (cfa-ribC) exhibiting overlapping TTS. Standard RNA-seq data track is shown in green for comparison. (inset) Predicted secondary structure for the overlapping region. Data are representative of three independent experiments. b, SEnd-seq data track and predicted secondary structure of an example overlapping TTS between a coding gene (sppA; red reads) and a non-coding antisense RNA (blue reads). Data are representative of three independent experiments. c, Venn diagram showing the number of overlapping bidirectional terminators identified for log versus stationary phase E. coli cells. d, Pie chart showing the fraction of overlapping TTS located between a gene pair or between a gene and an antisense ncRNA. e, (left) Average termination efficiency for all identified overlapping bidirectional terminators in either orientation (positive direction in red; negative direction in blue). n = 399. (right) Average termination efficiency for those bidirectional TTS that are located between a pair of highly expressed genes. n = 78. Error bars denote s.d. Data are representative of two independent experiments. f-i, Distributions of the length (f), folding energy (g), predicted stem size (h) and loop size (i) for the overlapping TTS. j, (left) Schematic of the stem-loop structure formed in the overlapping region. (right) Nucleotide profiles for the 5’ and 3’ flanking sequences of the stem-loop within an overlapping region. Such profiling allows for classification of the overlapping TTS into three categories. k, Pie chart showing the fraction of each category described in (j).