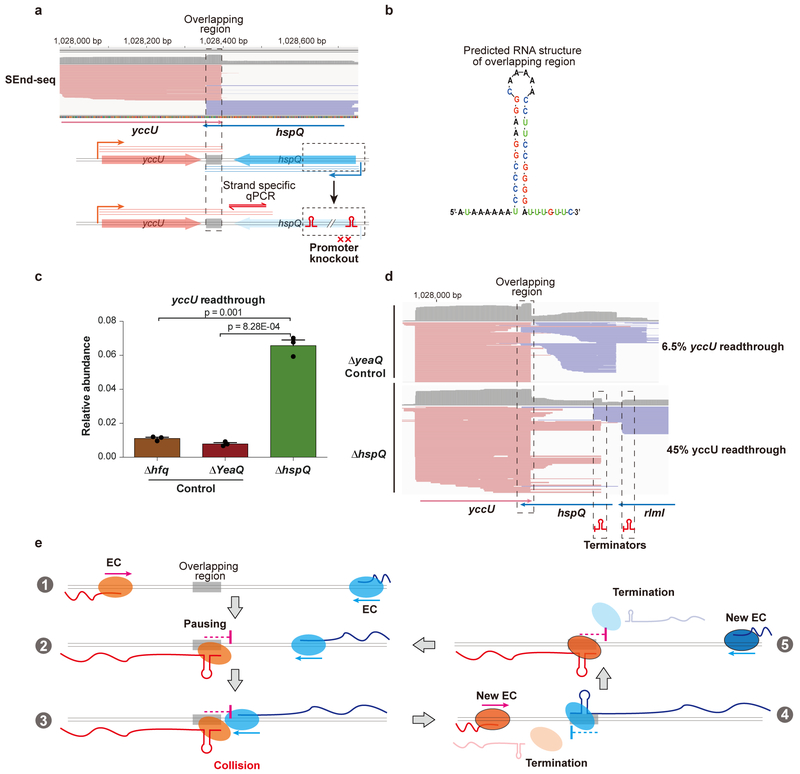
Fig. 6 |. Convergent transcription contributes to bidirectional termination in vivo.
a, SEnd-seq data track (top) and schematic of in vivo genomic modification (bottom) for the yccU-hspQ convergent gene pair. To disrupt hspQ transcription, we replaced the promoter and part of the gene body of hspQ with two strong intrinsic terminators. Data are representative of three independent experiments. b, Predicted secondary structure for the overlapping TTS between yccU and hspQ. c, qPCR results showing the relative abundance of yccU readthrough transcripts across the overlapping region when hspQ transcription is abolished (ΔhspQ). We also edited genes outside the convergent pair with the same procedure (Δhfq and ΔyeaQ) as controls. Data are mean ± s.d. from three independent experiments. P values were determined by two-sided unpaired Student’s t-tests. d, SEnd-seq data track around the yccU-hspQ region for the ΔyeaQ (top) or ΔhspQ strain (bottom). The fraction of yccU readthrough transcripts for each strain is indicated. Data are representative of two independent experiments. e, Model illustrating that head-on collisions between converging RNA polymerases drive bidirectional termination. The overlapping region produces an RNA hairpin that traps the transcription machinery, which is dislodged by another elongation complex traveling from the opposite direction—either through direct physical interaction or via torsional stress accumulated in the DNA. This process occurs repeatedly, resulting in highly efficient termination in both directions.