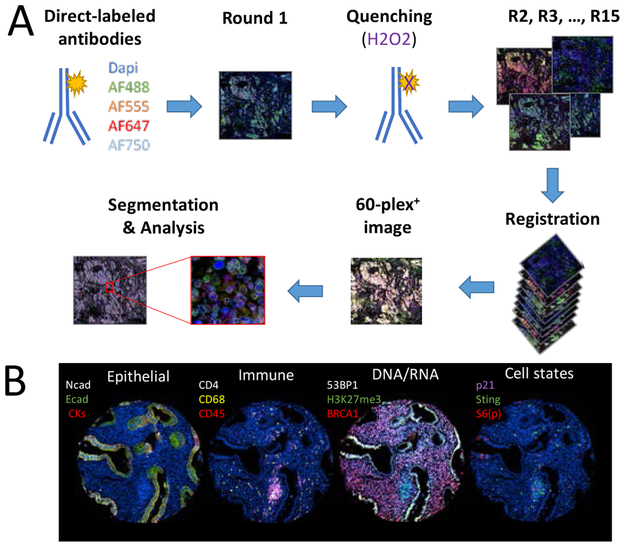
Figure 1. The use of cyclic-immunofluorescence for spatially oriented single cell proteomics.
(A)Primary antibodies are directly conjugated to fluorescent molecules. During the first round, fours antibodies conjugate with different Alexa-Fluor molecules are incubated on the tissue sample. Dapi is added to stain the cell nucleus and an image is acquired using a slide scanner. The signal is then quenched with hydrogen peroxide solution. Staining is then repeated for multiple rounds. The images are aligned during registration, which allows the creation of a multiplex image. Segmentation is then performed in order to analyze the expression and localization of each markers. (B) Example of a multiplex image acquired through cyclic immunofluorescence. Each panel represent a single region of a fallopian tube sample, with the fallopian tube being the source of ovarian cancer. In the first panel on the left, epithelial markers (N-cadherin, E-cadherin and a mix of cytokeratin-5, −7, −18 and −19) are being used to identify epithelial cells. In the second panel, immune cells are identified (CD68, CD4 and CD45). In the third panel, DNA and RNA related proteins (BRCA1, H3K27me3 and 53BP1) are stained. In the last panel on the right, cell states are identified (phosphorylated S6_ser235/ser236, p21 and STING).