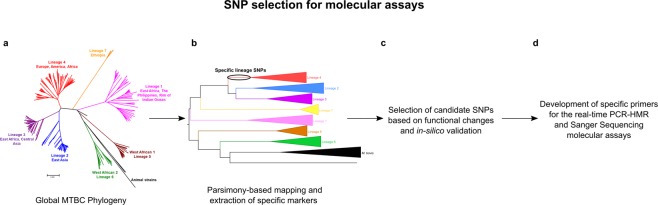
Figure 1.
Workflow to identify and select diagnostic lineage/sub-lineage SNP for the development of the molecular assays. (A) We used a publicly available variant list containing 34,167 SNPs from 219 MTBC global strains. (B) First, we performed a parsimony-based approach to map and obtain 2,056 specific SNPs markers for all lineages, and 1,337 for all L4 sub-lineages. In the figure, the specific SNPs for lineage 4 in the phylogeny are marked with a circle. (C) Then, candidate SNPs were selected according to their functional change (we used synonymous SNPs in essential genes, whenever possible), and tested in-silico against a global dataset of 4,495 MTCB strains. (D) Finally, we developed seven specific primers to implement in the real-time PCR-HRM molecular assay, and two genomic regions for Sanger sequencing approach.