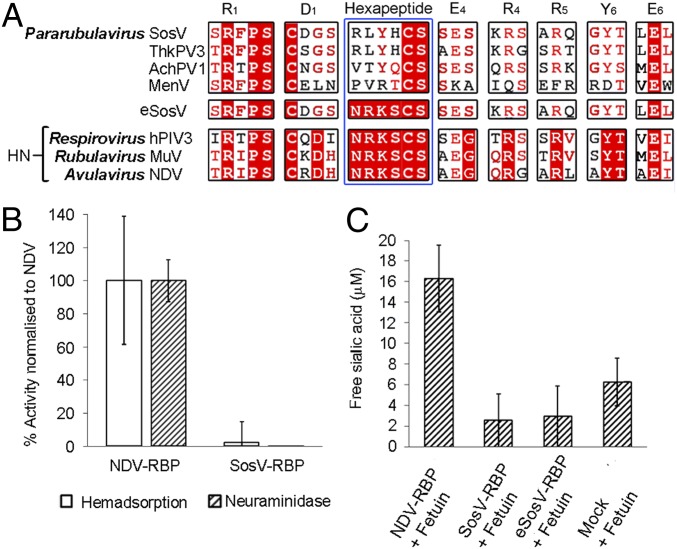
Fig. 1.
Amino acid sequence alignment and functional analysis indicate that SosV likely uses a sialic acid-independent mode of entry. (A) Aligment of the RBP amino acid sequences from SosV (YP_009094033.1), Tuhokovirus 3 (ThkPV-3) (YP_009094079.1), Achimota virus 1 (AchPV1) (YP_009094457.1), Menangle virus (MenV) (AAK62284.1), human parainfluenza virus 3 (hPIV3) (AAP35240.1), mumps virus (MuV) (BAA76983.1), Newcastle disease virus (NDV) (Q9Q2W5.1), and a construct of SosV engineered to incorporate the full hexapeptide motif (termed eSosV). The seven conserved sialidase residues (35, 37) and hexapeptide motif (40) are labeled according to residue and blade location (35) and annotated above the alignments. (B) SosV-RBP neuraminidase (48) and hemadsorption (47) activity normalized to cell surface expression and a NDV-RBP control. (C) Free sialic acid concentration detected following incubation of NDV-RBP, SosV-RBP, “NRKS” mutant eSosV-RBP, and mock-transfected cell supernatant with fetuin (49). For B (n = 10) and C (n = 6), error bars represent the SD.