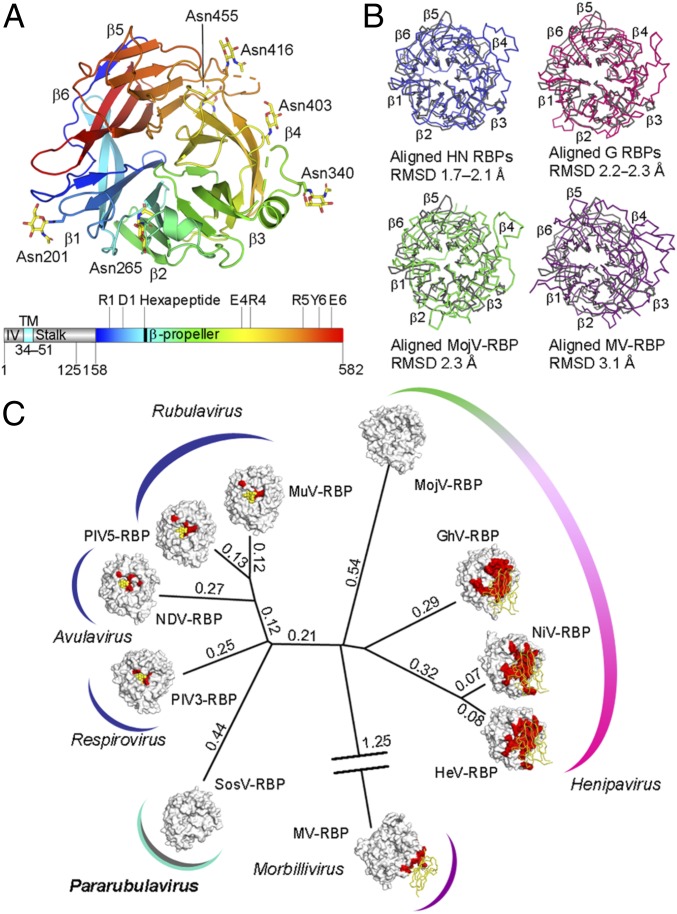
Fig. 2.
Structural relationship of SosV-RBP with other paramyxoviral RBPs. (A) Structure of a SosV-RBP β-propeller domain with propellers labeled (colored blue to red from N to C terminus, cartoon representation). Crystallographically observed N-linked glycosylation is represented as yellow sticks. Gene diagram with the predicted intraviral region (IV), transmembrane domain (TM), stalk, and β-propeller region annotated (colored as above). The sialidase residues and hexapeptide motif are annotated. (B) Overlays of SosV-RBP (gray) with other paramyxoviral RBP structures: clockwise from Upper Left, NDV, Newcastle disease virus (blue, 1E8V) (36); NiV, Nipah virus (pink, 2VWD) (94); MV, measles virus (2ZB5) (21); MojV, Mojiang virus (green, 5NOP) (62). Cα trace rendered and RMSD annotated. (C) Structure-based phylogenetic analysis of paramyxoviral RBP monomers: SosV, Sosuga virus; PIV3, parainfluenzavirus 3 (1V2I) (63); NDV (1E8V) (36); PIV5, parainfluenza virus 5 (4JF7) (64); MuV, mumps virus (5B2C) (43); MojV (5NOP) (62); GhV, Ghana virus (4UF7) (61); NiV (2VWD) (94); HeV, Hendra virus (2X9M) (22); MV (2ZB5) (21). Evolutionary distance matrices were calculated through pairwise superposition of RBP structures using SHP (60), and the unrooted tree was plotted in PHYLIP (92). RBPs are shown with surface representation. Relevant receptors are represented using ribbon (protein) or sphere (carbohydrate) (yellow). Known receptor binding sites are colored red on the glycoprotein surfaces. Calculated structure-based evolutionary distances are indicated beside the branches.