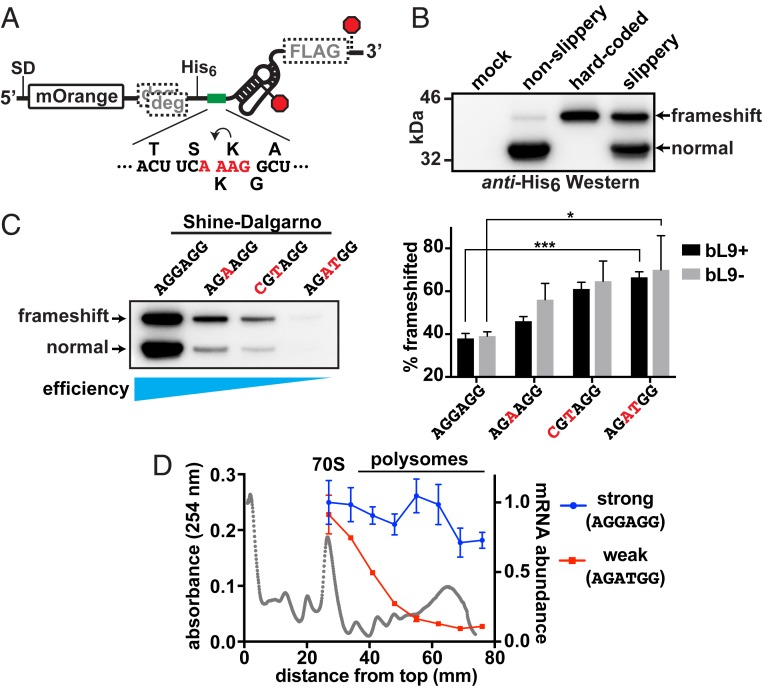
Fig. 1.
Ribosome abundance on an mRNA influences frameshifting. A reporter construct based on the IS3 pseudoknot was used to evaluate the impact of ribosome load on −1 frameshifting. (A) Schematic of the reporter showing the location of the SD sequence that was altered to adjust translation initiation efficiency, the mOrange2 protein domain, out-of-frame ClpXP degrons (gray “deg”), a His6 tag, a test sequence (green bar), the IS3 pseudoknot stimulator, and a FLAG tag in the −1 frame. Stop codons for the 0 and −1 reading frames are marked with red octagons. The test sequence is displayed below, with the encoded amino acids in the 0 (top) and −1 (bottom) frames and slippery tetrad nucleotides in red. (B) Anti-His6 Western blot revealing the products produced from a mock culture, a nonslippery reporter version lacking the slippery tetrad (A_AAG to C_AAG), a hard-coded “frameshift” product, and a slippery reporter with a strong consensus SD sequence. The canonical normal product and −1 frameshifted products are indicated. (C) (Left) Western blot of reporters with SD sequences altered to reduce translation initiation. As the translation initiation efficiency waned, the percentage of frameshifted product increased. (Right) Bar graph of frameshifted product percentages from separate quantitative Westerns of 3 biological replicates. Error bars are 1 SD. Asterisks signify t test P values < 0.05 (*) and <0.001 (***). (D) Sucrose gradients of lysates prepared from cells that had expressed the reporters with the strongest and weakest SD were used to separate ribosome forms prior to extracting RNA for reverse-transcription RT-qPCR. An absorbance profile is shown in gray with the locations of the 70S monosomes and polysome peaks. Primers that amplified the 5′ ends of the reporter mRNAs and 16S rRNA were used to establish the message/ribosome ratios, which were normalized to the amount of strong mRNA observed in the 70S peak. Error bars represent 1 SD of 3 experimental replicates.