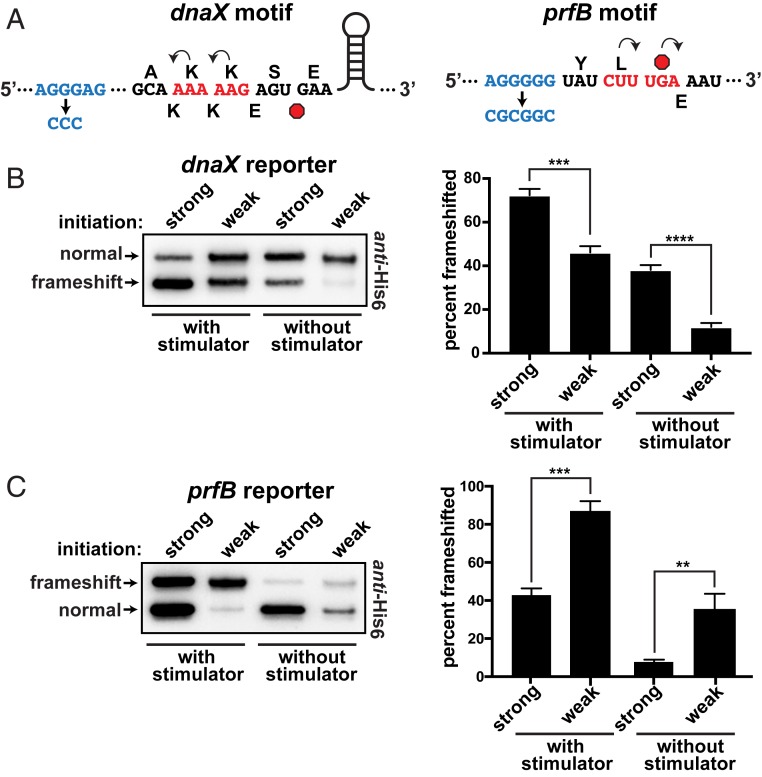
Fig. 2.
Frameshifting at the dnaX and prfB motifs. (A) Schematics of the dnaX and prfB programmed frameshift motifs that were cloned into the reporter system. The upstream SD-like stimulators are shown in blue, with their deactivated versions below. The 0-frame amino acids are shown above and the frameshifted amino acids are shown below each sequence. Stop codons are red octagons. The dnaX motif also contains a downstream stem loop stimulator that was not altered. Note that, for the dnaX motif, −1 frameshifting produced the smaller translation product. (B) Frameshifting at the dnaX motif as a function of ribosome load. The construct with a strong SD sequence (AGGAGG) exhibited more frameshifting than a version with a weak SD sequence (AGATGG). Inactivation of the nearby SD-like stimulator reduced frameshifting and increased the influence of ribosome load. (Left) To even the abundances for presentation clarity, the anti-His6 Western had the strong samples diluted 35- and 25-fold for the with stimulator and without stimulator samples, respectively. (Right) Separate Westerns of 3 biological replicates were used to generate the data for the bar chart. (C) Frameshifting at the prfB motif. The construct with a strong SD sequence (AGGAGG) exhibited less frameshifting than the version with a weak SD sequence (AGAAGG); (Left) the strong versions were diluted 10-fold for the presented Western. Inactivation of the nearby SD-like stimulator reduced frameshifting and increased the influence of ribosome load. (Right) Separate Western blots of 3 biological replicates were used to generate the data for the bar chart. Error bars represent 1 SD. Asterisks indicate t test P values of <0.01 (**), <0.001 (***), and <0.0001 (****).