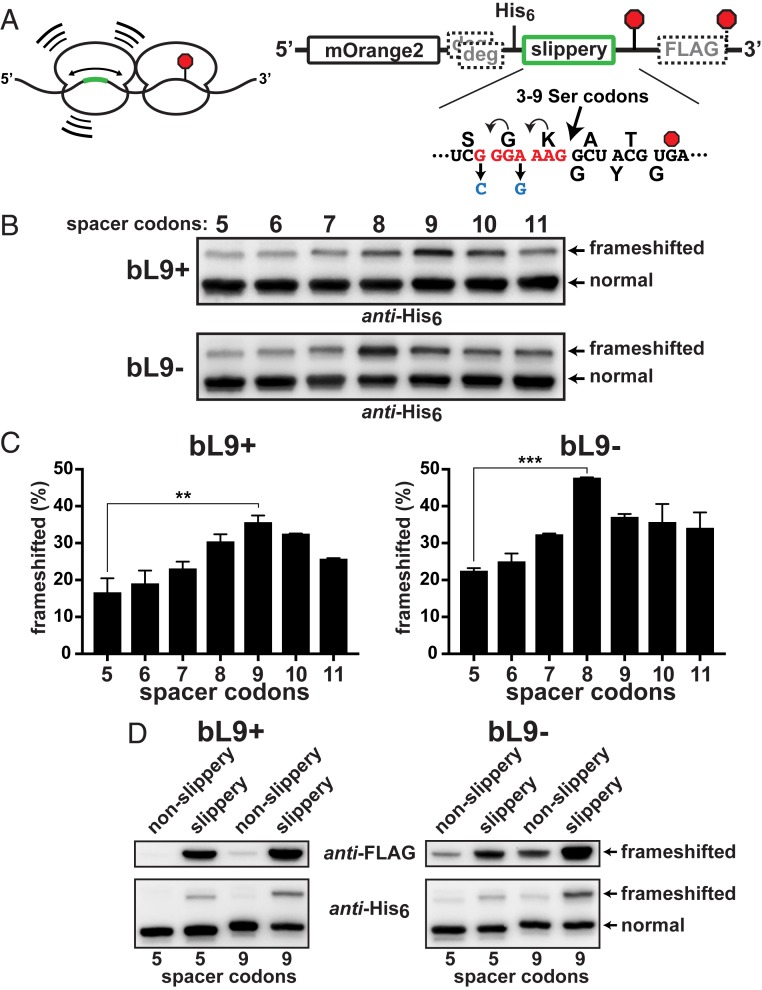
Fig. 4.
Stalled ribosomes are frameshift stimulators. A series of reporters was constructed to evaluate the influence of ribosome collisions on −1 frameshifting. (A, Left) Schematic of a collision between a ribosome transiently stalled at a stop codon (red octagon) and a trailing ribosome engaged with a slippery sequence located at different upstream positions (green). (Right) A detailed schematic. A slippery heptad sequence was positioned at varying distances 5′ to a slowly decoded UGA stop codon (red octagon). Nonslippery versions contained changes (blue) that hampered repositioning of the tRNAs. A downstream FLAG-tag appendage was encoded in the −1 frame. Out-of-frame ClpXP degrons preceded the test regions. The distance between the slippery sequence and the stalling stop codon was varied by inserting additional Ser codons such that the final A-site to A-site spacing ranged from 5 to 11 codons. (B) Anti-His6 Western blots of the reporter series that was expressed in bL9+ and bL9− cells. Frameshifting was evident in each case, but peaked with a 9-codon spacer in bL9+ and with an 8-codon spacer in bL9− cells. (C) Quantifications from 2 biological replicates were averaged and plotted. The peak frameshifting levels were significantly higher than the levels with the 5-codon spacer, and the bL9− cells exhibited more frameshifting overall. P values < 0.01(**) and <0.001 (***) are indicated. (D) (Top) Anti-FLAG and (Bottom) anti-His6 Westerns showing comparisons between reporters with 5 or 9 spacer codons with and without the slippery heptad.