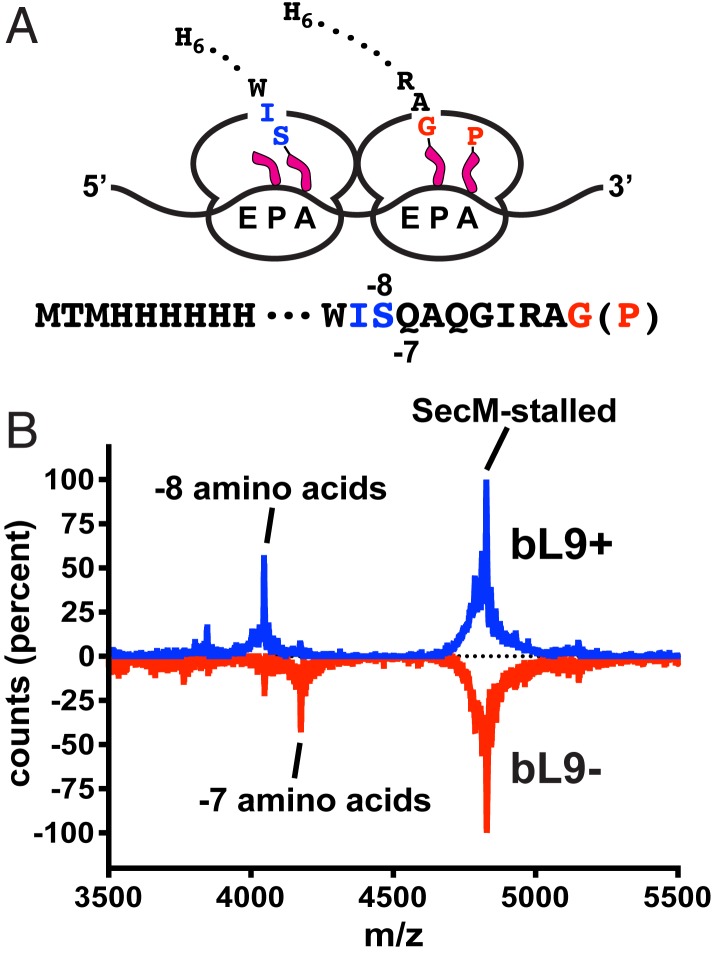
Fig. 5.
Ribosomal protein bL9 alters interribosomal spacing. A construct was designed to stably stall ribosomes so that the positions of trailing ribosomes could be established by characterizing their nascent peptides. (A) Schematic of 2 colliding ribosomes and the nascent peptide sequences. The lead ribosome is stably engaged with a SecM stalling motif with peptide-tRNAGly in the P-site and tRNAPro in the A-site (red). The colliding ribosome contains a shorter nascent peptide, with deacylated tRNAIle in the P/E-site and peptide-tRNASer in the A/P-site (blue). The peptides contained N-terminal His6 tags that allowed for purification under denaturing conditions prior to mass spectroscopy. (B) MALDI-TOF mass spectra of peptides recovered from sucrose gradient polysome fractions of bL9+ (blue) and bL9− cells (red, inverted). The dominant mass for the SecM-stalled polysomes corresponded to a peptide ending at the Gly residue of the SecM sequence shown in A (theoretical m/z = 4,801.40 Da, observed = 4,801.14 Da). Peptides recovered from bL9+ polysomes also contained a prominent mass corresponding to a peptide that was 8 residues shorter than the SecM-stalled peptide (theoretical = 4,019.53 Da, observed = 4,019.42). The peptides from bL9− polysomes contained an additional peptide 7 residues shorter (theoretical = 4,147.66 Da, observed = 4,146.90 Da).