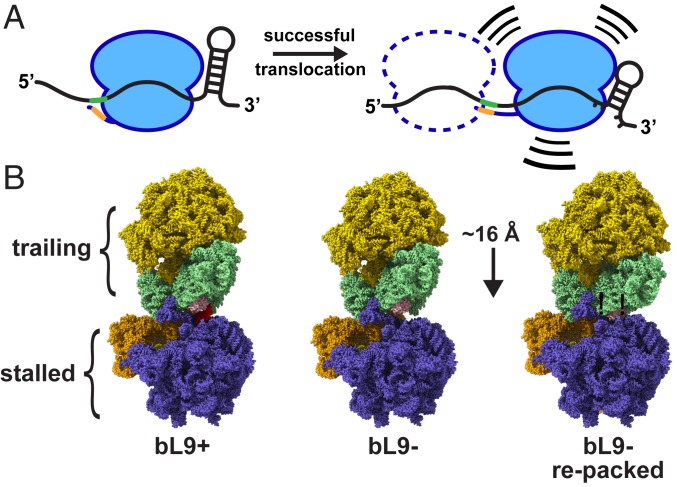
Fig. 8.
Models of mRNA tension and collisions. (A) Schematic of a ribosome in 2 locations relative to a frameshift element containing flanking stimulators. (Left) The SD sequence in the mRNA (green) interacts with the anti-SD (orange) in a relaxed manner, and the downstream mRNA stem loop is relaxed. (Right) A ribosome that has translocated farther into the motif, with a tensioned SD:anti-SD structure and a partially unfolded stem loop. The combination of the 2 flanking forces provides the energy to increase −1 frameshifting. A potential trailing ribosome is depicted as a dashed line. The trailing ribosome may promote entry of the lead ribosome into a compromised location while still allowing for a −1 frameshift. (B) Ribosome collision models with a stalled ribosome and a trailing ribosome. (Left) A cryo-EM structure of a ribosome awaiting translation termination (stalled, PDB ID 6ore; slate and orange) that is docked to a structure of a ribosome in the resting state (trailing, PDB 6osq; gold and light green) using a collision orientation similar to that observed in many crystal structures. Protein bL9 of the stalled ribosome is colored red, and protein uS4 of the trailing ribosome is colored pink. (Center) Protein bL9 was removed while maintaining the orientations of both particles. (Right) The trailing ribosome was reoriented and moved closer such that uS4 of the trailing ribosome accommodated the space previously occupied by bL9. This relocation reduced the intervening mRNA length by approximately one codon.