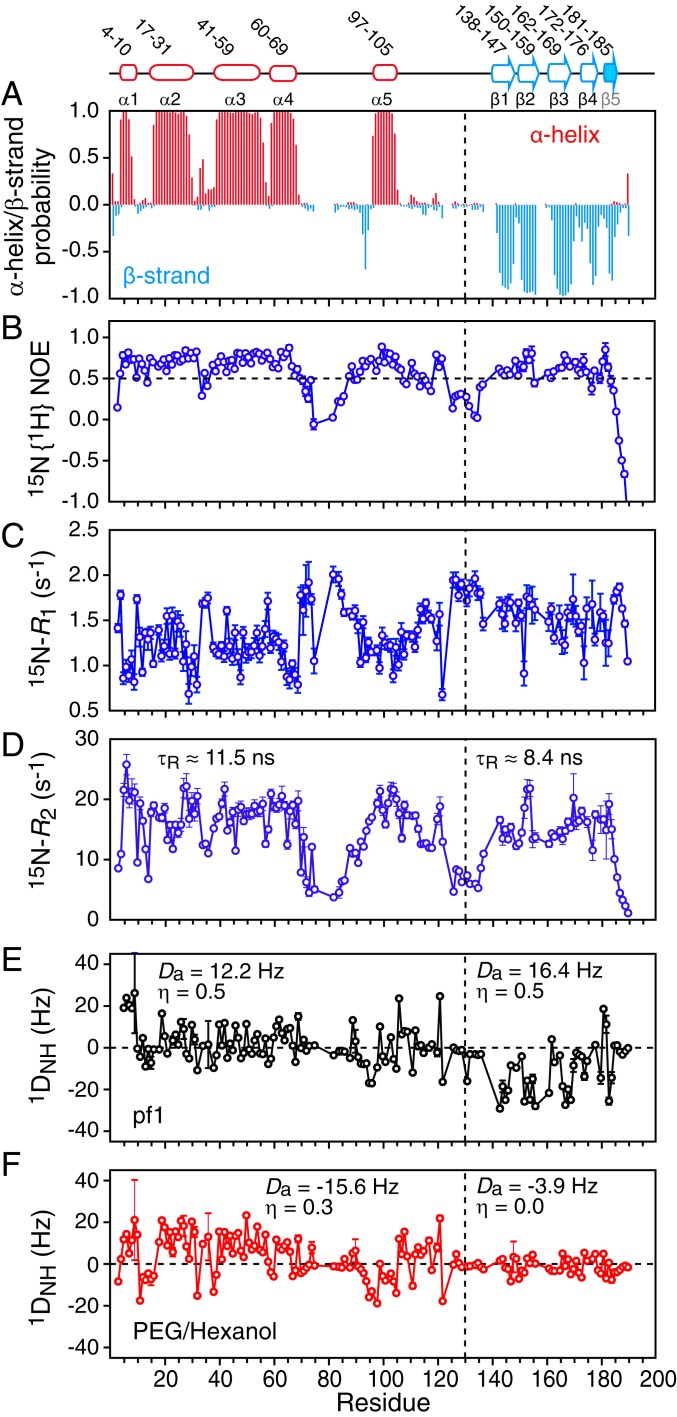
Fig. 2.
NMR characterization of ΔST-DNAJB6b. (A) TALOS-derived secondary structure (α helix in red and β strand in cyan) probability derived from the backbone 13Cα, 13Cβ, 13C′, 15N, and 1HN chemical shifts. The 15N relaxation data: (B) 15N{1H} heteronuclear NOE, (C) 15N-R1, and (D) 15N-R2. The 15N-R2 values were calculated from 15N-R1ρ values recorded with a 1.5-kHz spin lock field after correction for 15N-R1. Backbone amide (1DNH) residual dipolar coupling (RDC) data collected in 2 different alignment media: (E) 15 mg/mL phage pf1 and (F) 4.2% (vol/vol) polyethylene glycol/hexanol. The values of the magnitude of the principal component of the alignment tensor (Da) and the rhombicity (η) were determined from the distribution of RDCs using a maximum likelihood method (35, 36). The relaxation and RDC data were recorded at 600 and 800 MHz, respectively, on 120 μΜ 2H/15N-labeled ΔST-DNAJB6b samples at 25 °C.