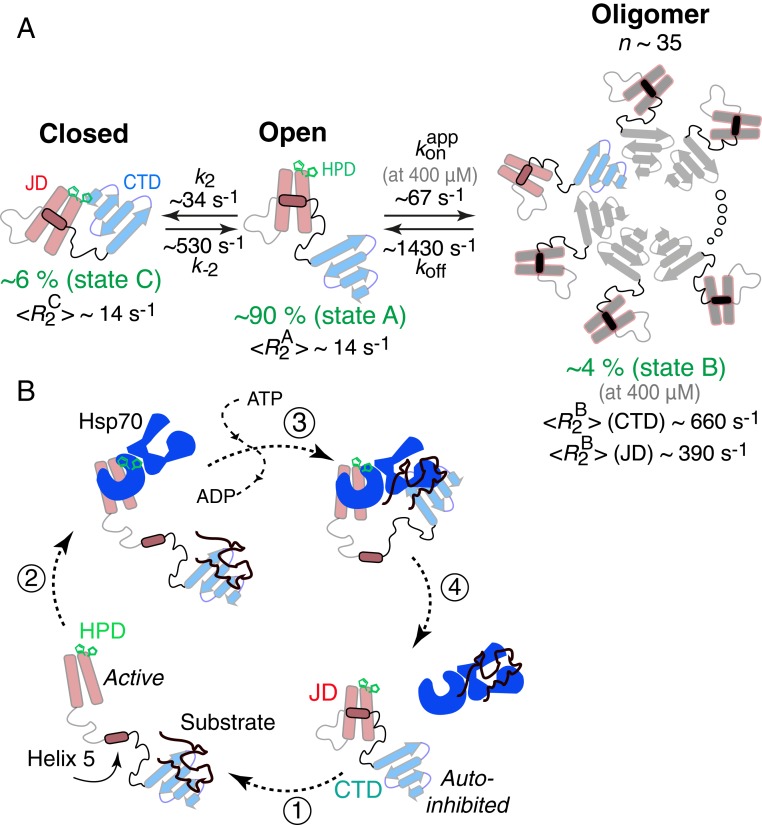
Fig. 8.
Kinetic scheme used to fit the 15N relaxation-based NMR data for ΔST-DNAJB6b and proposed mechanism of Hsp70 (DnaK) cycle regulation by Hsp40 (DnaJ). (A) The scheme involves a major open species (state A; pA ∼ 90%) in exchange with a closed monomeric species (state B; pB ∼ 6%), in which the HPD sequence motif plays an important role, and a large oligomeric state (state C; pC ∼ 4% at 400 μΜ). Based on the fitted 15N- values (at 25 °C) for the CTD domain, one can estimate that the oligomer is ∼850 kDa in molecular weight and consists of ∼35 monomeric units. (B) Proposed Hsp40 (DnaJ)–Hsp70 (DnaK) cycle. In the absence of a substrate, Hsp40 exists in an autoinhibited state where helix 5 (containing the DI/VF sequence motif) blocks the Hsp70 binding site. Upon substrate binding (step 1), a conformational change occurs that exposes the Hsp70 binding site allowing Hsp70 to bind via the HPD sequence motif (step 2). After ATP hydrolysis and substrate handover (step 3), the cycle is completed by the displacement of the Hsp70 ATPase domain by helix 5 of Hsp40 (step 4). Although DNAJB6b exists as an oligomer in vivo, we only show the monomer as this proposed mechanism is expected to be valid for other DnaJ proteins.