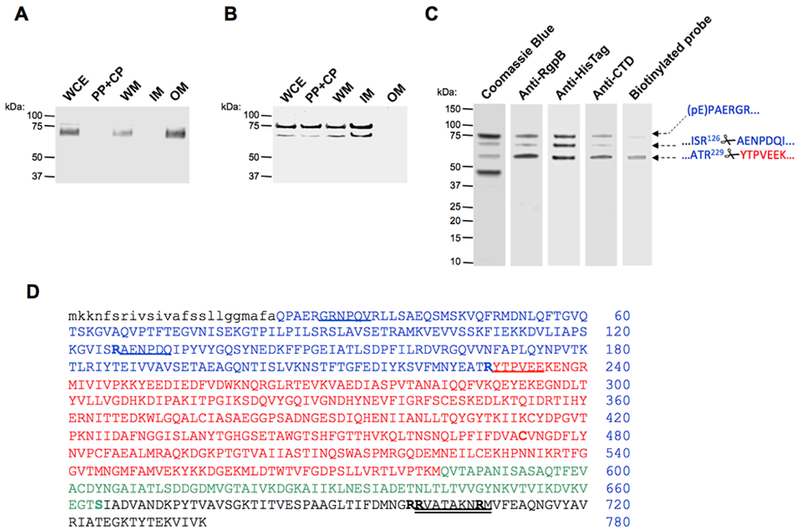
Figure 1. Subcellular location of proRgpB6His produced in wild-type P. gingivalis (panel A) and the T9SS-deficient ΔporN mutant (panel B).
(A & B) Whole cells extracts (WCE) were fractionated into the following fractions: periplasm and cytoplasm (PP + CP), whole membrane (WM), inner-membrane (IM) and outer-membrane (OM). The localization of proRgpB was determined by Western blotting using anti-RgpB polyclonal antibody. (C) ProRgpB6His from the ΔPorN mutant strain was purified by affinity chromatography on Ni-Sepharose and final products were analyzed by SDS-PAGE and Western blotting with anti-RgpB, anti-6His and anti-RgpBCTD antibodies, and by active-site labeling using the specific biotinylated probe, BiRK. N-terminal amino acid sequences of proteins indicated by arrows were determined by automated Edman-degradation and mass spectrometry analysis. (D) The amino acid sequence of preproRgpB with individual domains coded: lowercase: signal sequence; BLUE: N-terminal pro-domain (PD); RED: catalytic domain; GREEN: Ig-like domain; BLACK: C-terminal domain (CTD). R126 and R229 of the pro-domain and sequences identified by the Edman degradation of indicated (arrows) bands are underlined. Double underlined is a RgpBCTD-derived peptide used to obtain rabbit polyclonal anti-RgpBCTD. Residues mutated in the CTD; to Arg (S664) and to Lys (R700, R701 and R708) are highlighted (see Fig 6).