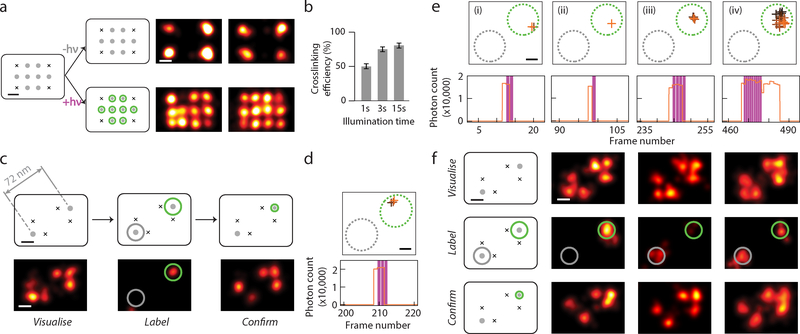
Fig. 2. Crosslinking efficiency test and targeted single-molecule labelling.
(a) Test of CNVK labelling strand crosslinking efficiency on single molecules. Left, DNA nanostructure design with four corner markers and eight central crosslinking targets, spaced 20 nm point-to-point, before and after crosslinking. Right, DNA-PAINT images after crosslinking. Top, negative control, bottom, with 405 nm laser illumination.
(b) Crosslinking efficiency comparison for different illumination time. Error bars represent Poisson counting uncertainty (total number of grids counted: 103, 60, 82, respectively, each having 8 binding sites; number of labelled targets: 419, 349, 510, respectively).
(c) Schematics and representative images at different stages of targeted single-molecule labelling experiment. Top row, schematics of DNA test structure design with two candidate targets, spaced 72 nm apart; bottom row, super-resolution images. Within bottom row, left panel shows pre-acquisition image of candidate target sites together with markers, middle panel shows real-time labelling session image of CNVK labelling strand transient blinking events, and right panel shows post-acquisition image of labelled targets together with markers.
(d,e) Examples of real-time blinking and laser activation traces, comprising of (d) a single blinking event and laser pulse for the sample in (c), and (e) a series of four events for the sample in (F), left column. For each detected blinking event, XY traces (top) and time traces (bottom) are shown.
(f) Representative examples of successful labelling experiments. Top row, pre-acquisition images; middle row, real-time labelling session images; bottom row, post-acquisition images.
In (a,c,f), grey dots without and with green halo indicate idle and labelled targets, black crosses indicate fixed position markers; green and grey circles overlaid on super-resolution images indicate desired and undesired target sites, respectively.
In (d,e), XY traces are colour-coded by time (from black to orange) and overlaid with include- (green, dotted circle) and exclude- (grey, dotted circle) ROIs; time traces show detected photon count per single-molecule localisation (orange), overlaid with crosslinking laser illumination events (magenta).
See Supplementary Fig. 8–9 for additional representative examples of both successful and unsuccessful single-target labelling experiments, and Supplementary Notes 3.4 for discussion regarding statistics and reproducibility.
All scale bars, 20 nm.