Figure 1.
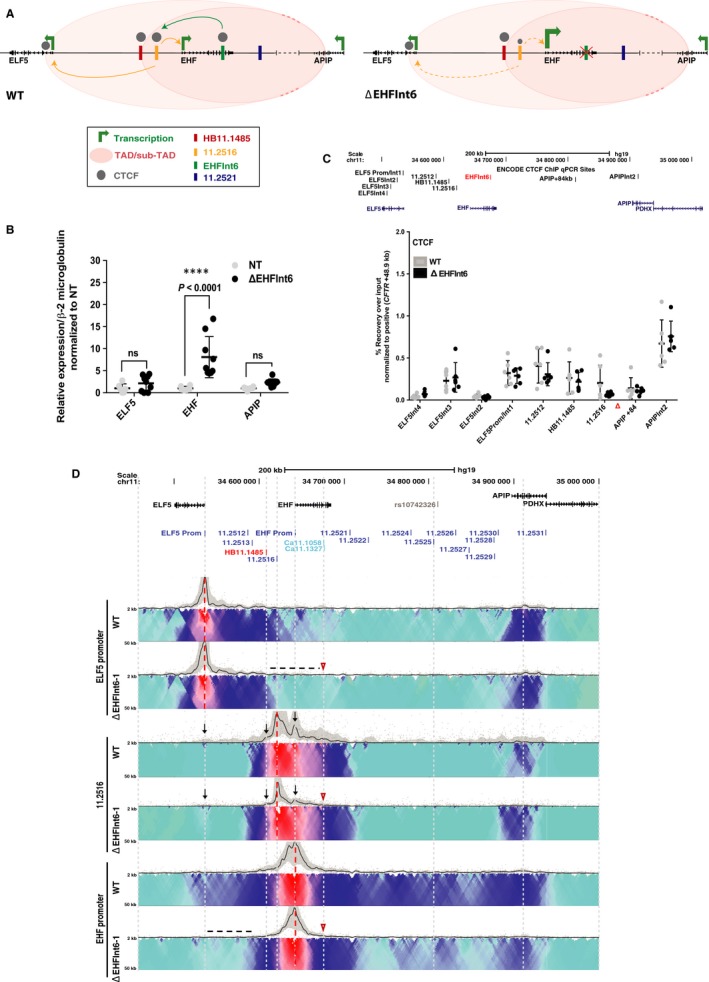
Impact of deletion of cis‐element EHF intron 6 in A549 c9 cells. (A) EHF intron 6 maintains a structural role at 11p13 and helps to facilitate looping of the 11.2516 enhancer (green arrow) to the ELF5 and EHF promoters (yellow arrows). Upon deletion of EHF intron 6 (red X), EHF expression is enhanced. Additionally, CTCF occupancy at 11.2516 is reduced (smaller grey circle) and its interactions with the ELF5 and EHF promoters are weakened (yellow dashed arrows). (B) RT‐qPCR for ELF5, EHF and APIP expression in non‐targeted WT (NT) and deletion clones. Data are normalized to β2M and relative to clonal WT. Error bars are standard deviation (SD), n ≥ 3. ****P < .0001, ***P < .001, **P < .01, *P < .05, ns = not significant by a two‐way ANOVA test with multiple comparisons. (C) ChIP‐qPCR for CTCF enrichment at interacting sites across the 11p13 region in WT and deletion clones. ChIP results are shown as percent recovery over input and are normalized to a positive control (CFTR + 48.9 kb (28). Error bars are SD, n ≥ 2. (D) Circular chromosome conformation capture (4C‐seq) data in WT and one EHF intron 6 deletion clone. This deletion removes the Ca11.1058 and Ca11.1327 DHS. Viewpoints at the ELF5 promoter, 11.2516 and EHF promoters are shown (red dotted lines). Open red arrowheads indicate the deletion site. The upper panel (black line) shows the main trend of interaction, and the colour‐coded domainogram indicates relative interactions with a window size ranging from 2‐50kb. Red denotes the strongest interactions; dark blue, to turquoise, to grey represent decreasing interaction frequencies. Informative interactions or loss thereof are shown as black dotted lines or black arrows
