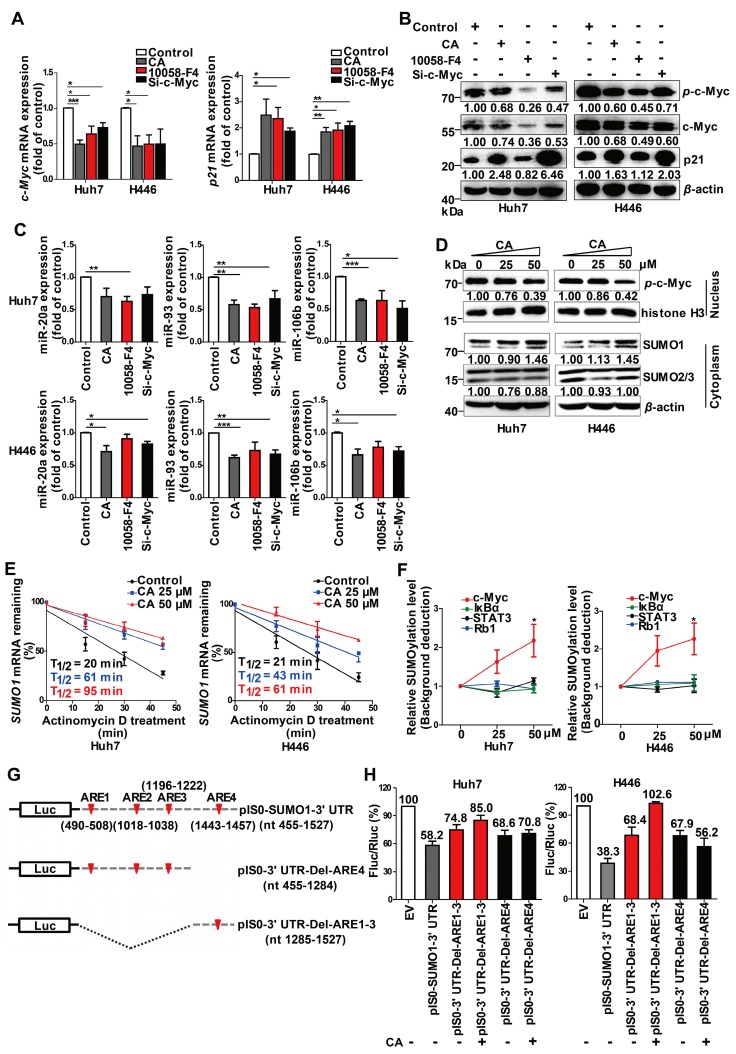
Figure 4.
SUMOylation of c-Myc by CA decreased p-c-Myc and miR-17 family and increased p21. Huh7 and H446 cells were treated with CA (50 μM), or 10058-F4 (30 μM, c-Myc-specific inhibitor), or c-Myc siRNA (50 nM), respectively, for 24 h. (A) Expression of c-Myc (left) and p21 (right) mRNAs by qRT-PCR assay. (B) Protein expression of p-c-Myc, total c-Myc, and p21 in Huh7 (left) and H446 (right) cells by Western blotting; the results were normalized to β-actin in density scanning with the control group as 1. (C) Levels of the miR-17 members in Huh7 (upper) and H446 (lower) cells, treated with CA or 10058-F4 or c-Myc siRNA at conditions mentioned above, assessed by qRT-PCR. (D) Expression of nuclear protein p-c-Myc in Huh7 (left) and H446 (right) cells by Western blotting and normalized to histone H3 with density scanning (upper); Cytoplasmic proteins SUMO1 and SUMO2/3 in Huh7 and H446 cells determined by Western blotting and normalized to β-actin (lower), with the control (0 μM) as 1. (E) CA increased SUMO1 mRNA stability. Huh7 (left) and H446 (right) cells were treated or untreated with CA for 12 h. Subsequently, actinomycin D (5 μM) was added at indicated time points. mRNA degradation was analyzed by qRT-PCR. Decay curves were plotted versus time. (F) Sumoylation of c-Myc, IκBα, Rb1, and STAT3 in Huh7 (left) and H446 (right) cells treated with CA (25, 50 μM,) was analyzed using the EpiQuikTM Protein Sumoylation Assay Kit (Epigentek). (G) Schematic presentation of the SUMO1 mRNA 3' UTR inserted into pIS0 vector and construction of Luc-SUMO1 mRNA 3' UTR fusions. (H) ARE-4 region involved in the stabilizing effect of CA on SUMO1 mRNA, using dual luciferase reporter assay. Data were normalized to EV as 100%. EV stands for empty vector of pIS0. All data are presented as mean ± SEM of 3 independent experiments. * p < 0.05, ** p < 0.01 and *** p < 0.001 (treated vs untreated control).