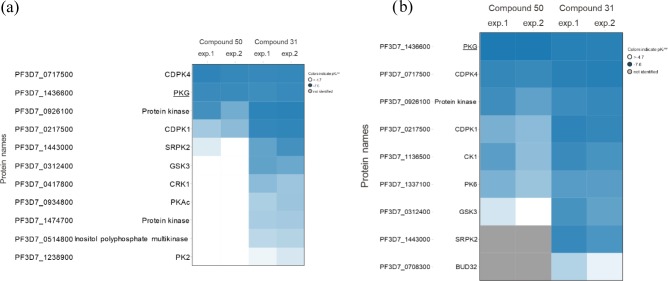
Figure 4.
Chemoproteomics profiling of compounds 31 and 50. (a) Both compounds were profiled on Kinobeads, which represent a combination of immobilized promiscuous ATP-competitive kinase inhibitors, in a P. falciparum protein extract. A total of 54 P. falciparum kinases were analyzed. The concentration of the “free” compounds used for competition, compound 50 and compound 31, was between 0.08 and 20 μM over six samples. (b) Compounds 50 and 31 were profiled with a bead matrix generated by immobilizing either compound 50 or compound 31 to beads, and competed with the respective “free” analogue over six concentrations between 0.08 and 20 μM. The Heatmaps show the protein kinases affected by any of the two compounds in two independent experiments, respectively. The values shown are apparent pKd values (blue: decreasing apparent pKd values; white: no competition; gray: protein not identified).