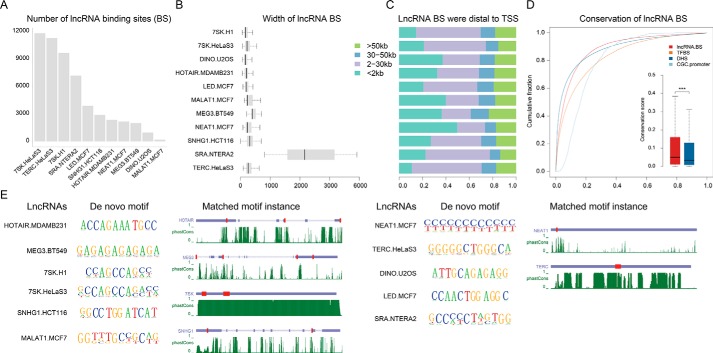
Figure 1.
Systematic identification of lncRNA-chromatin–binding sites. A, the statistic of lncRNA- binding sites identified in human. B, lncRNA-binding sites in human were focal. C, most of the lncRNA-binding sites were distal to its closest genes. The binding sites occurred more than 2 kb from TSS were considered as distal. D, cumulative distributions of sequence conservation scores of lncRNA-binding sites, DHS, transcription factor-binding sites (TFBS), and promoters of CGC genes. The significance test was performed using a Wilcoxon rank-sum test (***, p value < 0.001; n.s., not significant, p value > 0.05). E, motif discovery and scanning of lncRNA-binding sites in human using HOMER and FIMO, respectively. For motif discovery, only the motif with the highest significant level was shown. For motif scanning, only the top 3 motif instance ranked by conservation were shown in the transcripts. The red boxes represented the matched motif instances in lncRNAs. The green lines represent conservation calculated by phastCons.