ABSTRACT
We uncovered a role for nucleoli and PML-bodies as phase-separated protein quality control organelles that compartmentalize protein quality control factors and misfolded proteins for their efficient clearance. Failure to dispose misfolded proteins converts nucleoli and PML-bodies into a solid state that immobilizes ubiquitin, limiting its recycling for genome integrity maintenance.
KEYWORDS: Protein quality control, nucleus, membraneless organelles, genome instability
Author’s comment
Eukaryotic cells are constantly exposed to a multitude of stressors that challenge their proteome, thus threatening cell functionality. Maintenance of nuclear protein homeostasis (or proteostasis) is particularly important to preserve genome integrity. Imbalances in nuclear proteostasis are linked to age-related neurodegenerative diseases, many of which are characterized by nuclear protein aggregates and genomic instability .1 Although nuclear protein aggregation induces mutagenesis and DNA damage, we do not understand how these processes are linked on the molecular level. Moreover, we still have a limited understanding of how the nucleus copes with protein misfolding.
Maintenance of the nuclear proteome requires the transport of proteins through nuclear pores. This transport process is highly selective, although proteins with low molecular weight (< 40 kDa) can passively diffuse through these pores. In addition to regulating protein and RNA transport between the nucleus and cytoplasm, nuclear pores and the nuclear envelope also protect the nuclear proteome from misfolded proteins. The main source of misfolded proteins in mammalian cells are newly synthesized polypeptides that failed to reach their native state and defective ribosomal products (DRiPs), which cannot properly fold due to errors occurring during DNA replication (DNA mutations), transcription (damaged mRNAs) or translation (amino acid misincorporation, premature translation termination or pervasive translation outside of protein-coding regions) .2,3 Upon their synthesis in the cytoplasm, DRiPs are recognized and bound by the chaperones Heat Shock Protein Family A (HSP70) and Valosin Containing Protein (VCP), which assist their targeting for proteasome-mediated degradation or autophagy .1,4 Failure to clear DRiPs, due to HSP70 and VCP inhibition, leads to proteostasis imbalances and the aberrant accumulation of DRiPs inside ribonucleoprotein (RNP) granules .5 We recently showed that the accumulation of misfolded proteins in cytoplasmic membraneless organelles such as stress-inducible RNP granules causes their conversion into an aggregate-like state with pathological implications .6
In Mediani et al., we now show for the first time that DRiPs not only affect membraneless organelles in the cytoplasm, but also in the nucleus. We provide evidence that a fraction of low molecular weight DRiPs can diffuse into the nucleus and accumulate in two nuclear membraneless compartments: nucleoli and Promyelocytic Leukemia Protein (PML) bodies. Nucleoli and PML-bodies function, respectively, as constitutive and stress-responsive overflow compartments for DRiPs, which are then cleared by chaperones and proteasomes. The finding that defective proteins such as DRiPs are transiently stored inside the nucleolus of normally growing mammalian cells was surprising and highlights two important concepts: first, nuclear proteostasis is constantly challenged by the influx of low molecular weight misfolding-prone proteins; second, nuclear quality control compartments exert a buffering function by capturing DRiPs and preventing their aberrant interactions with other essential nuclear components (Figure 1).
Figure 1.
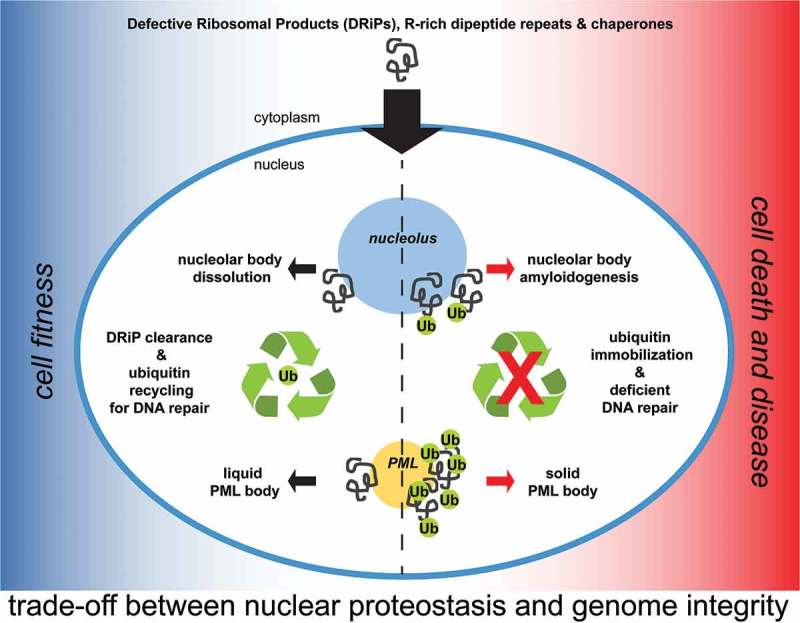
Role of nucleoli and PML-bodies in nuclear proteostasis. A fraction of low molecular weight DRiPs escapes the cytoplasmic protein quality control system and enters the nucleus. DRiPs are transiently stored in nucleolar subcompartments (NoABs) for later disposal by chaperones and proteasomes. Upon proteotoxic stress conditions, such as heat shock, and proteasome inhibition, a fraction of DRiPs is transiently compartmentalized at PML-bodies, which act as stress-responsive overflow compartments. Along with DRiPs, PML-bodies recruit ubiquitin, chaperones and proteasomes to promote efficient DRiP degradation. Failure to clear DRiPs from NoABs and PML-bodies leads to their solidification and immobilizes ubiquitin and proteasomes. This, in turn, depletes the pool of free ubiquitin to the detriment of the ubiquitin-dependent labeling of DNA damaged sites.
The accumulation of proteins inside nucleoli is not unprecedented. Previous studies identified nucleolar subcompartments that accumulate proteins upon stress conditions: the amyloid-body (A-body) and the nucleolar aggresome (NoA) .7,8 In addition, the granular subcompartment of the nucleolus was recently identified as a storage site for later refolding or disposal of misfolded proteins, including thermolabile firefly luciferase .9 Our study demonstrates that these nucleolar subcompartments are the same structure, referred to as Nucleolar A-bodies (NoABs), and establishes a new function for the nucleolus as protein quality control (PQC) compartment. The peptides that are stored inside NoABs seem to be heterogeneous: besides DRiPs, the misfolded model substrates firefly luciferase and VHL, as well as arginine-rich dipeptide repeats, which are associated with Amyotrophic Lateral Sclerosis (ALS), were transiently retained within NoABs .9 These misfolded proteins are cleared with the assistance of HSP70 and proteasomes. But what happens if DRiPs cannot be cleared from nucleoli? We found that if not efficiently removed by chaperones and proteasomes, DRiPs trigger amyloidogenesis of nucleoli, with potential consequences on nucleolar functions. Thus, irreversible NoAB aggregation may represent an important mechanism contributing to aging and disease-associated toxicity.
Some proteotoxic stress conditions, such as temperature upshift or proteasome inhibition, cause a strong increase in the total amount of DRiPs. When the DRiP concentration reaches a critical threshold in the nucleoplasm, PML-bodies are induced and act as stress-responsive compartments that concentrate DRiPs together with ubiquitin, proteasomes and chaperones (Figure 1). Thus, PML-bodies are stress-adaptable compartments that fulfill different functions in resting and stressed cells: in resting cells, PML-bodies function as post-translational and epigenetic control sites, whereas in stressed cells they are PQC and sorting sites for the efficient clearance of DRiPs. Similar to NoABs, failure to clear DRiPs from PML-bodies, due to chaperone impairment, convert them into an amyloid-like solid state that immobilizes ubiquitin and proteasomes (Figure 1).
Which are the functional consequences of sequestering large amounts of quality control factors in quality control compartments? Ubiquitin is not only required for PQC, but also to repair damaged DNA, making the nuclear ubiquitin levels a limiting factor under certain conditions. Moreover, proliferation of mammalian cells often goes along with some degree of DNA replication failure, leading to spontaneous DNA lesions. Considering that the repair of these lesions requires ubiquitination, 10 we investigated whether there is a trade-off between nuclear proteostasis and genome integrity. Indeed, we found that nuclear proteostasis and DNA repair compete for a limiting pool of ubiquitin. Importantly, upon proteotoxic stress conditions, ubiquitin is primarily used to label DRiPs for clearance, to the detriment of its use for DNA repair mediated by Tumor Protein P53 Binding Protein 1 (also known as 53BP1). Based on these findings, we conclude that NoAB and PML-body formation represents a protective stress response aimed at prioritizing the restoration of nuclear proteostasis. However, this comes at a price, because prolonged immobilization of ubiquitin and proteasomes at PML-bodies endangers genome stability and cell health. This suggests that severe stress conditions and chronic diseases that impair PQC function, such as polyglutamine diseases and ALS, also jeopardize genome stability and cell viability.
Together, these findings make a strong case for further investigations aimed at elucidating the quality control functions of nucleoli and PML-bodies and how dysregulation of these compartments affects genome integrity and contributes to human aging and disease. Recent studies highlighted that a substantial fraction of nascent peptides is destroyed within minutes after their synthesis by proteasomes. Some of these peptides are generated from upstream open-reading frames3 and globally these peptides are referred to as the “dark proteome”, being their sequence and structure unknown. This “dark proteome”, located in the cytoplasm and the nucleus, would challenge nuclear proteostasis by potentially competing with mutated and unstable proteins for folding and clearance. Its identification will uncover important molecular targets, whose handling may offer therapeutic perspectives for the treatment of protein conformation diseases. On the other hand, combined treatments with drugs that impair nucleoli and PML-body quality control functions may sensitize and efficiently kill cancer cells, which are characterized by high translation rates and genome instability.
Funding Statement
S.C. acknowledges AriSLA Foundation (Granulopathy and MLOpathy); Cariplo Foundation (Rif. 2014-0703); MAECI (Dissolve_ALS); and MIUR (Departments of excellence 2018–2022; E91I18001480001). S.C. and S.A. acknowledge EU Joint Programme—Neurodegenerative Disease Research (JPND) project. The project is supported through funding organizations under the aegis of JPND (http://www.neurodegenerationresearch.eu/). This project has received funding from the European Union’s Horizon 2020 Research and Innovation Programme under grant agreement No 643417. S.A. acknowledges the Max Planck Society, the ERC (no. 725836), and the BMBF (01ED1601A, 031A359A).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Klaips CL, Jayaraj GG, Hartl FU.. Pathways of cellular proteostasis in aging and disease. J Cell Biol. 2018;217(1):51–63. doi: 10.1083/jcb.201709072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schubert U, Antón LC, Gibbs J, Norbury CC, Yewdell JW, Bennink JR.. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404(6779):770–774. doi: 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- 3.Ingolia NT, Brar GA, Stern-Ginossar N, Harris MS, Talhouarne GJS, Jackson SE, Wills MR, Weissman JS. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep. 2014;8(5):1365–1379. doi: 10.1016/j.celrep.2014.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verma R, Oania RS, Kolawa NJ, Deshaies RJ. Cdc48/p97 promotes degradation of aberrant nascent polypeptides bound to the ribosome. Elife. 2013;2:e00308. doi: 10.7554/eLife.00308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ganassi M, Mateju D, Bigi I, Mediani L, Poser I, Lee HO, Seguin SJ, Morelli FF, Vinet J, Leo G, et al. A surveillance function of the HSPB8-BAG3-HSP70 chaperone complex ensures stress granule integrity and dynamism. Mol Cell. 2016;63(5):796–810. doi: 10.1016/j.molcel.2016.07.021. [DOI] [PubMed] [Google Scholar]
- 6.Zhang P, Fan B, Yang P, Temirov J, Messing J, Kim HJ, Taylor JP. Chronic optogenetic induction of stress granules is cytotoxic and reveals the evolution of ALS-FTD pathology. Elife. 2019;8. doi: 10.7554/eLife.39578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Audas TE, Audas DE, Jacob MD, Ho JJD, Khacho M, Wang M, Perera JK, Gardiner C, Bennett CA, Head T, et al. Adaptation to stressors by systemic protein amyloidogenesis. Dev Cell. 2016;39(2):155–168. doi: 10.1016/j.devcel.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Latonen L, Moore HM, Bai B, Jäämaa S, Laiho M. Proteasome inhibitors induce nucleolar aggregation of proteasome target proteins and polyadenylated RNA by altering ubiquitin availability. Oncogene. 2011;30(7):790–805. doi: 10.1038/onc.2010.469 [DOI] [PubMed] [Google Scholar]
- 9.Frottin F, Schueder F, Tiwary S, Gupta R, Körner R, Schlichthaerle T, Cox J, Jungmann R, Hartl FU, Hipp MS. The nucleolus functions as a phase-separated protein quality control compartment. Science. 2019;365:342–347. doi: 10.1126/science.aaw9157. [DOI] [PubMed] [Google Scholar]
- 10.Lukas C, Savic V, Bekker-Jensen S, Doil C, Neumann B, Pedersen RS, Grøfte M, Chan KL, Hickson ID, Bartek J, et al. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat Cell Biol. 2011;13(3):243–253. doi: 10.1038/ncb2201. [DOI] [PubMed] [Google Scholar]


