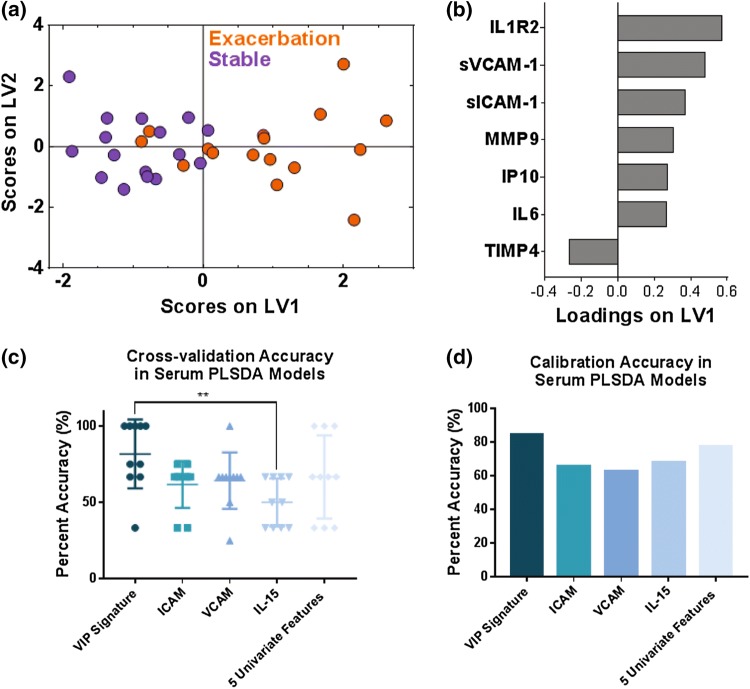
Figure 2.
VIP scores and PLSDA identified a signature of 7 serum proteins that differentiated stable from exacerbation measurements in 16 paired stable and AE-COPD events experienced by 11 unique patients. (a) VIP scores identified a 7-protein serum signature that differentiated stable (purple) and exacerbation (orange) events with 81.25% cross-validation accuracy and 84.38% calibration accuracy. Latent variable 1 (LV1) accounted for 25.00% of the variance in the data, and latent variable 2 (LV2) accounted for 16.75% of the variance in the data. (b) The loadings plot shows how much each protein contributes to the signature, with positive loadings associated with exacerbation events, and negative loadings comparatively reduced in exacerbation. (c) Comparison of the differentiation between stable and exacerbated states based on individual factors vs. multivariate signatures. The VIP signature identified by the PLSDA models trended towards higher cross-validation accuracy than individual factors that were most significantly different. A one-way ANOVA determined that this signature was significantly better than IL-15 alone, with ** indicating a p value less than 0.01 after Tukey’s test for multiple comparisons. (d) Comparison of the calibration accuracies for individual factors vs. the VIP signature identified by the PLSDA model.