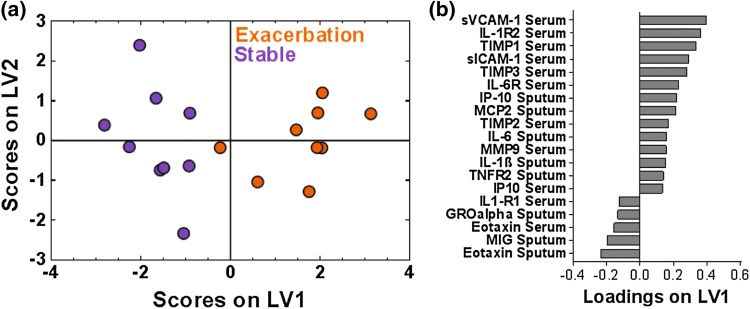
Figure 3.
A one latent variable PLSDA model of VIP-selected proteins from the combined serum and sputum samples combined resulted in clear differentiation between stable and exacerbation measurements across 9 paired stable and AE-COPD events experienced by 7 unique patients. (a) PLSDA and VIP scores identified a signature of 19 proteins that differentiated the stable (purple) from exacerbation (orange) states with 88.89% cross-validation and calibration accuracy. Latent variable 1 accounted for 21.73% of the variance in the data. The scores plot shown is based on a two latent variable model to enable better visualization of group separation. (b) The loadings plot illustrates the protein contributions to the VIP-selected signature, with positive loadings positively associated with the exacerbation measurements, and negative loadings comparatively reduced during exacerbation.