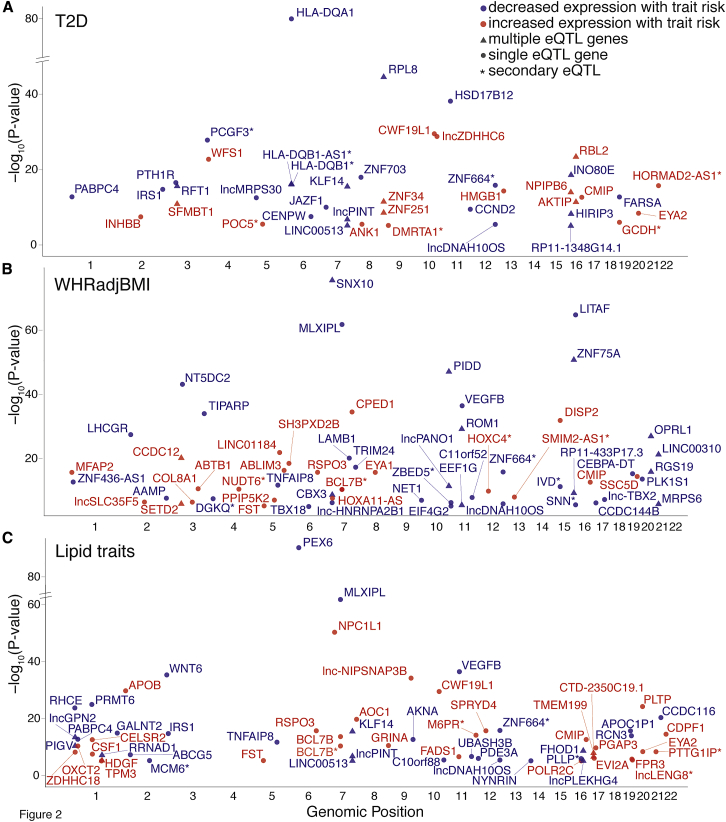
Figure 2.
Cardiometabolic GWAS and eQTL Colocalizations, Stratified by Trait
Each point represents the lead GWAS variant of a signal colocalized by LD and conditional analysis with an eQTL for the named gene. Plots show eQTLs at GWAS loci for (A) T2D, (B) WHRadjBMI, and (C) lipids. The x-axis shows chromosomal positions of colocalized signals and the y-axis shows the −log10 p values of the GWAS variant’s association with gene expression level in adipose tissue. After matching the effect allele from the eQTL study with the risk allele from the GWAS, risk alleles that are associated with increased gene expression level are shown in red and risk alleles that are associated with decreased gene expression level are shown in blue. Triangles indicate that more than one gene is colocalized with the GWAS variant (other genes would appear at the same x-axis position). Variants colocalized as secondary eQTLs are designated by a star after the gene name. Only the strongest associations for the named traits are shown; full results can be found in Table S9.