Figure 1.
APC2 Bi-allelic Loss of Function Mutations in Posterior-Predominant (P > A) Lissencephaly
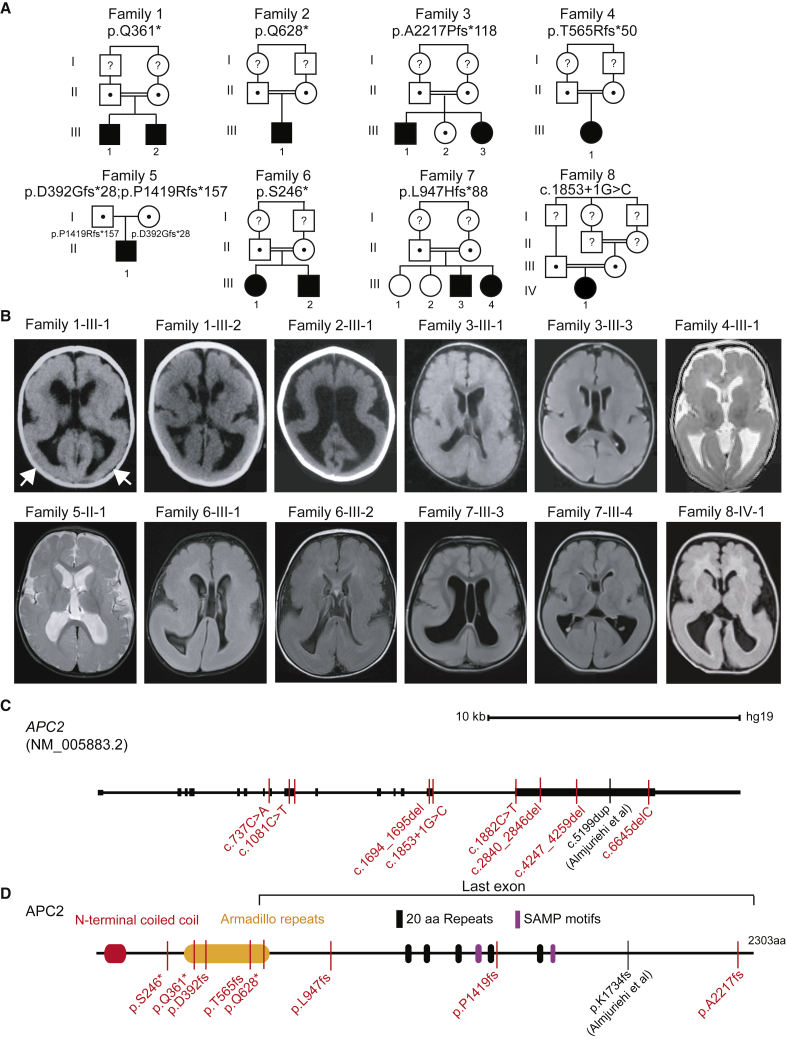
(A) Twelve affected individuals from eight families showed unique bi-allelic mutations in APC2. All families except family 5 had documented parental consanguinity (double bars). The allele is listed below the family number. Dots and question marks indicate heterozygous carriers and samples not tested, respectively.
(B) Axial brain imaging in each family showed evidence of posterior-predominant lissencephaly. In families 1 and 2, only brain CT was available, but for other families, brain MRI is shown. Scans showed more severe agyria in the posterior than in the anterior cortex (arrows in first scan highlight posterior agyria). Scans are T1 or FLAIR sequences, except for those of families 4 and 5, which are T2 sequences.
(C) Gene organization of APC2. The scale bar represents 10 kb. APC2 contains 15 exons, the first of which is non-coding. Mutations were scattered throughout the coding region of the protein; five mutations were present in large exon 15. Exon 15 also contains the homozygous c.5199dup Almuriekhi et al. mutation.16
(D) APC2 is a 2,303 amino acid multidomain scaffolding protein containing an N-terminal coiled-coil, Armadillo repeats, 20 amino acid (aa) repeats (FXVEXTPXCFSRXSSLSSLS), and SAMP (Ser-Ala-Met-Pro) motifs. Affected individuals’ mutations, represented with simplified nomenclature, were located throughout the open reading frame. The region of the protein encoded by the last exon is highlighted.