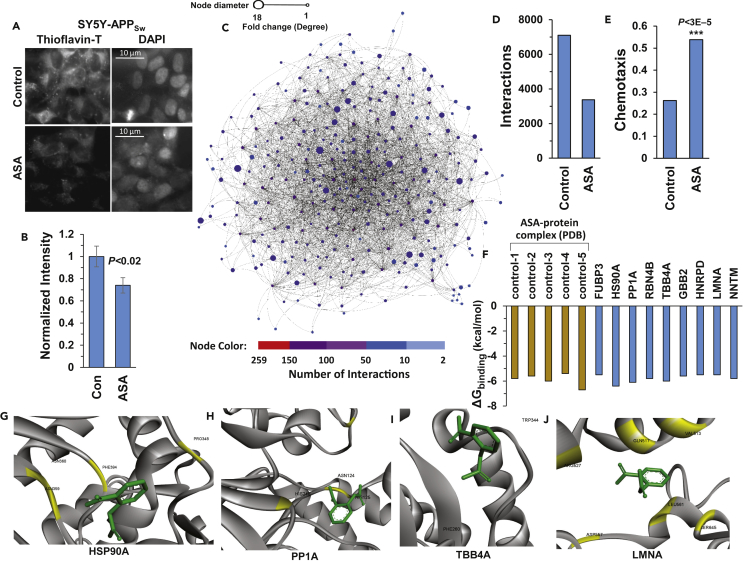
Figure 6.
Aspirin Treatment Reduces Aggregate Complexity
(A) Thioflavin-T staining of amyloid in SY5Y-APPSw cells (images on left), ± simultaneous exposure to 0.5 mM aspirin. DAPI staining of nuclei (images on right) demonstrates similar cell density. Scale bar indicates 10 μm.
(B) Normalized quantitations of amyloid-like aggregation per cell in SY5Y-APPSw cells, with or without aspirin treatment. Data are shown as mean ± S.E.M. *p < 0.02 by one-tailed t test.
(C) The insoluble-aggregate interactome of SY5Y-APPSw cells exposed to 0.5 mM aspirin for 48 h shows substantially reduced hub degree and complexity relative to untreated cells (compare with Figure 1C). See also Figure S2.
(D) Number of aggregate-network interactions is reduced by half in SY5Y-APPSw cells exposed to 0.5 mM aspirin, relative to untreated control cells.
(E) Aspirin (1 mM) protects SY5Y-APPSw cells against chemotaxis decline in C. elegans strain CL2355 (pan-neuronal Aβ42 expression) relative to vehicle-only controls. ***p < 2.5 × 10−5, significance by chi-squared (χ2) test.
(F) Interaction energies (ΔGbinding) predicted by computational docking of aspirin with candidate proteins (blue bars, network-implicated hub proteins) versus previously reported ASA-binding targets (tan bars, five positive controls to confirm correct docking parameters).
(G–J) Aspirin docking poses of proteins predicted to be direct ASA-binding targets (Ayyadevara et al., 2017): (G) HSP90A (heat shock protein 90α); (H) PP1A (see Figure 5A legend); (I) TUBB4A (tubulin beta chain 4A, a major constituent of microtubules); and (J) LMNA (lamin 4 A/C, a constituent of nuclear lamina; mutations are implicated in several muscular and cardiac dystrophies and in Hutchinson-Gilford progeria).