Figure 1.
Profiling circRNA after Spliceosome Inhibition in Primary Rat Hippocampal Neurons
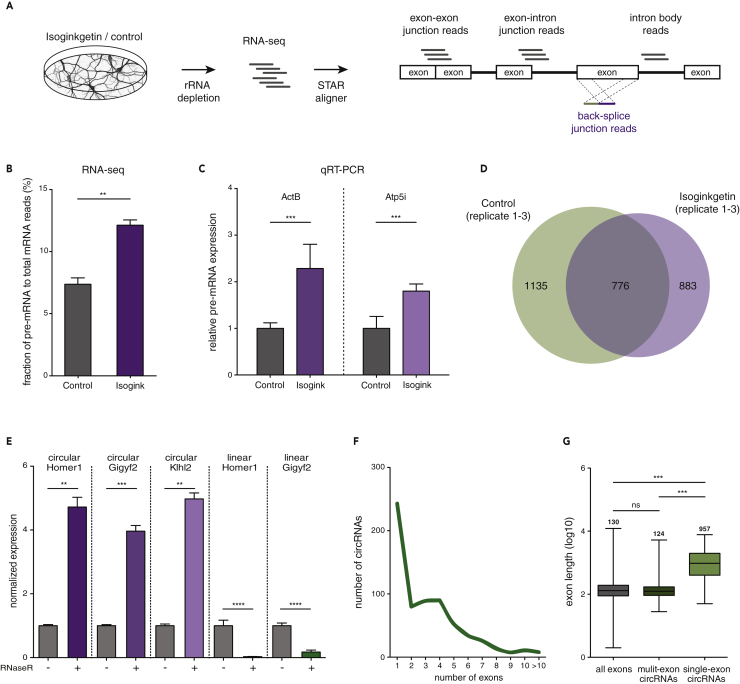
(A) Experiment and analysis pipeline.
(B) The abundance of pre-mRNA in a total mRNA sample was quantified after Isoginkgetin (purple) or control (gray) treatment. Assessed by RNA-seq, Isoginkgetin treatment led to a significant increase in the pre-mRNA population (**p < 0.01, unpaired t test, two-tailed, n = 3).
(C) Relative pre-mRNA expression for two candidate ActB and ATP5I transcripts under control and Isoginkgetin treatment conditions assessed with qRT-PCR (***p < 0.001, unpaired t test, two-tailed, n = 3).
(D) Number of circRNAs identified in all replicates in the Isoginkgetin and control conditions.
(E) Validation of circularity using RNase R treatment followed by qRT-PCR. CircRNAs increased in expression, whereas linear transcripts were depleted after RNase R incubation (****p < 0.0001, ***p < 0.001, **p < 0.01, unpaired t test, two tailed, n = 4).
(F) Numbers of exons that comprise the population of 776 circRNAs.
(G) Length distribution of back-spliced exons. A median length of 957 and 124 bp was found for single- and multi-exon circRNAs, respectively (***p < 0.0001, Kruskal-Wallis test followed by Dunn's multiple comparisons test). Error bars represent SD (B, C, and E). Box-whisker plots show minimum, first quartile, median, third quartile, and maximum of a set of data (G).