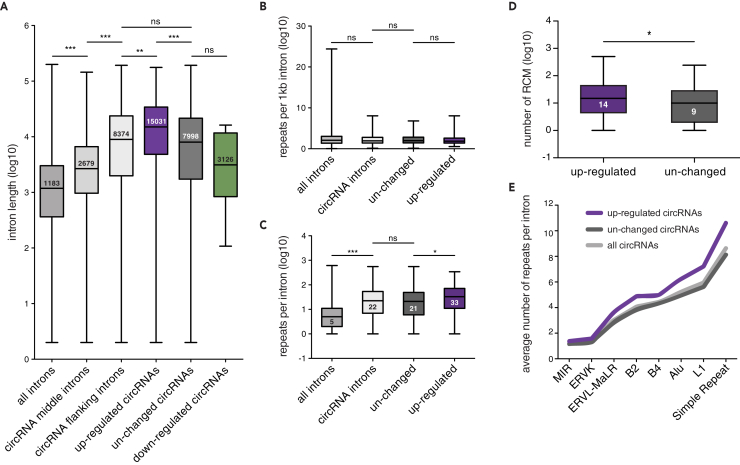
Figure 3.
Distinct Intronic Features Facilitate circRNA Formation under Conditions of Reduced Spliceosome Activity
(A) Length distribution of introns. Introns flanking back-spliced exons are significantly longer than the median rat intron and circRNA middle introns (***p < 0.0001, Kruskal-Wallis test followed by Dunn's multiple comparisons test). Flanking introns of up-regulated circRNAs are significantly longer than flanking introns of unchanged circRNAs and all circRNAs (***p < 0.0001, **p < 0.01, Kruskal-Wallis test followed by Dunn's multiple comparisons test).
(B) Frequency of repeat sequence occurrence per 1 kb. No difference is observed between all introns, all circRNA-flanking introns, flanking introns of unchanged circRNAs, and up-regulated circRNAs (ns > 0.05, Kruskal-Wallis test followed by Dunn's multiple comparisons test).
(C) Number of repeat sequences detected per flanking intron. Flanking introns of circRNAs harbor more repeats than the average rat intron. Up-regulated circRNAs contain significantly more repeat sequences than the flanking introns of unchanged circRNAs (***p < 0.0001, *p < 0.05, Kruskal-Wallis test followed by Dunn's multiple comparisons test).
(D) Number of Reverse complementary motifs (RCM) detected in flanking intron pairs. Flanking introns of up-regulated circRNAs possess significantly more RCMs compared with unchanged circRNAs (*p < 0.05, Mann Whitney U test).
(E) Comparison of the average number of repeats observed in the flanking intron. Distinct repeat families (x axis) are more abundant in flanking introns of up-regulated circRNAs compared with unchanged circRNAs, whereas the number of repeat families observed between all circRNAs and the unchanged population is similar. Box-whisker plots show minimum, first quartile, median, third quartile, and maximum of a set of data (A–D).