Figure 2.
Single-Nucleus RNA Sequencing of the Developing Human Kidney and Pseudotemporal Ordering Resolves Major Cell Types and Informs on Podocyte Developmental Programs
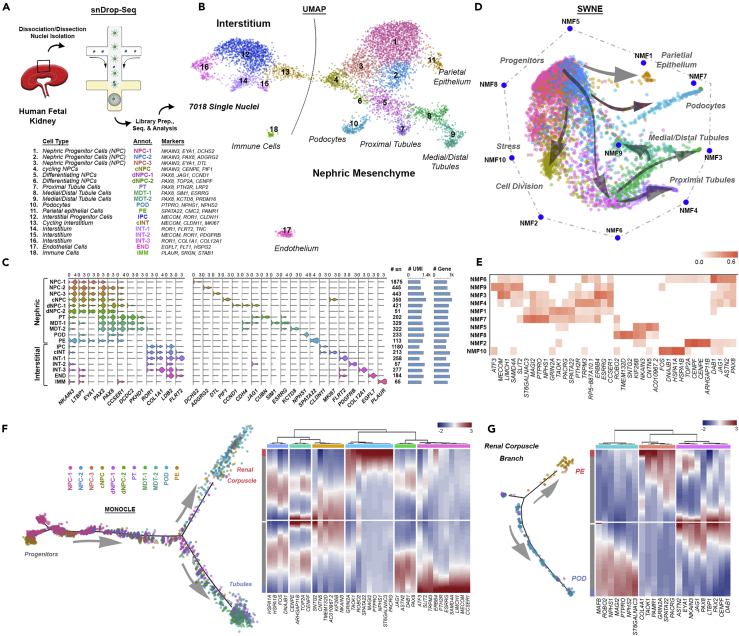
(A) An overview of the snDrop-seq pipeline on fetal kidney samples (Table S1) including dissociated kidney cortical cells (14 and 16 weeks) and sieved glomeruli (13 and 15 weeks).
(B) Combined expression data (four individuals over five experiments) visualized by UMAP dimensional reduction showing 18 distinct cellular populations encompassing the nephrogenic, interstitial, endothelial, and immune cell types. Cell type annotations and associated select marker genes are indicated for each cluster.
(C) Violin plots showing expression values for select broad or cell-type specific marker genes. For each cluster, the number of datasets (SN), average number of transcripts (UMI), and average number of genes detected are indicated.
(D) SWNE visualization of nephrogenic lineage cell clusters and associated non-negative factorization (NMF) identified gene signatures indicate cluster relationships.
(E) Heatmap of top genes associated with each NMF state.
(F) Trajectory analysis using Monocle of nephrogenic lineages showing developmental progression into distinct RC (proximal) and tubule (medial/distal) lineages. Corresponding heatmap indicating discrete gene expression intensity from the progenitor state (gray center line value) maturing to either RC (red tip value) or medial/distal (blue tip value) branches.
(G) Trajectory analysis of the developing RC branch into either parietal or visceral (podocyte) epithelial cell types. Heatmap showing expression values of select marker genes progressing from differentiating NPCs (center, gray) to either parietal (red) or podocyte (blue) cell types.
See also Figures S1–S4.