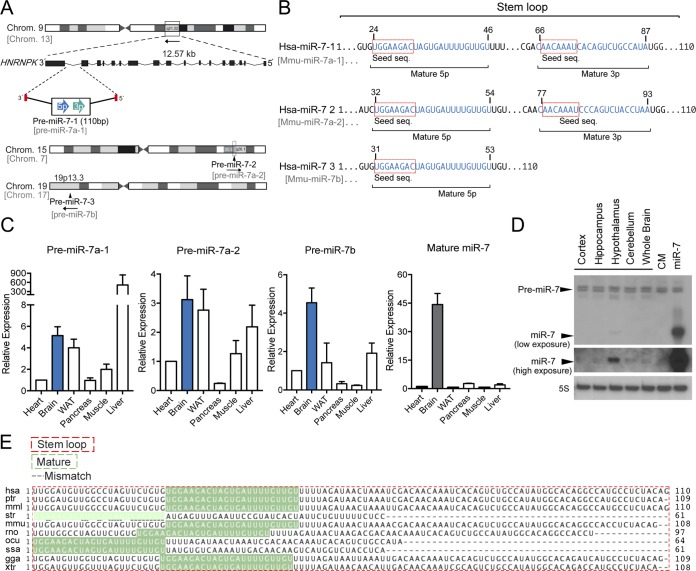
FIG 1.
miR-7 genomic locations, mouse tissue expression, and conservation of precursor and mature miR-7 among species. (A) Schematic representation of the genomic locations of miR-7 family members in human and mouse. Human miR-7-1 and murine miR-7a-1 are located in introns 16 and 17 of the HNRNPK gene, respectively. miR-7-2 and miR-7-3 are located in intergenic regions of human chromosomes 15 and 19. Murine miR-7 genes and their chromosome locations are shown in gray. (B) Sequence alignment between human miR-7 family members. Seed sequences are indicated in boxes. Blue indicates the mature 5p and 3p sequences. (C) Gene expression analyses (qRT-PCR) of precursors and mature miR-7-1-5p in different mouse tissues normalized to snoRD68. Data are expressed relative to the amount of miR-7-5p transcripts expressed in the heart. Results are means ± SEM from three independent experiments. (D) Representative Northern blot analysis of miR-7-5p in different brain tissues showing an enrichment in mouse hypothalamus. 5S rRNA was used as a loading control. (E) Sequence alignment of the stem-loop and mature miR-7 among species. WAT, white adipose tissue; hsa, Homo sapiens (human); ptr, Pan troglodytes (chimpanzee); mml, Macaca mulatta (rhesus macaque); str, Stronglyloide ratti; mmu, Mus musculus (mouse); rno, Rattus norvegicus (rat); ocu, Oryctolagus cuniculus (rabbit); ssa, Salmo salar (Atlantic salmon); gga, Gallus gallus (chicken); xtr, Xenopus tropicalis (western clawed frog).