Stress granules (SGs) are ribonucleoprotein aggregates that form in response to stress conditions. The regulation of SG dynamics is not fully understood. Permanent pathological SG-like structures were reported in neurodegenerative diseases such as amyotrophic lateral sclerosis. The Ras GTPase-activating protein-binding protein G3BP1 is a central regulator of SG dynamics. We found that the lysine 376 residue (K376) of G3BP1, which is in the RRM RNA binding domain, was acetylated.
KEYWORDS: acetylation, CBP/p300, G3BP1, HDAC6, stress granule, stress proteins, stress response
ABSTRACT
Stress granules (SGs) are ribonucleoprotein aggregates that form in response to stress conditions. The regulation of SG dynamics is not fully understood. Permanent pathological SG-like structures were reported in neurodegenerative diseases such as amyotrophic lateral sclerosis. The Ras GTPase-activating protein-binding protein G3BP1 is a central regulator of SG dynamics. We found that the lysine 376 residue (K376) of G3BP1, which is in the RRM RNA binding domain, was acetylated. Consequently, G3BP1 RNA binding was impaired by K376 acetylation. In addition, the acetylation-mimicking mutation K376Q impaired the RNA-dependent interaction of G3BP1 with poly(A)-binding protein 1 (PABP1), but its RNA-independent interactions with caprin-1 and USP10 were little affected. The formation of G3BP1 SGs depended on G3BP1 RNA binding; thus, replacement of endogenous G3BP1 with the K376Q mutant or the RNA binding-deficient F380L/F382L mutant interfered with SG formation. Significant G3BP1 K376 acetylation was detected during SG resolution, and K376-acetylated G3BP1 was seen outside SGs. G3BP1 acetylation is regulated by histone deacetylase 6 (HDAC6) and CBP/p300. Our data suggest that the acetylation of G3BP1 facilitates the disassembly of SGs, offering a potential avenue to mitigate hyperactive stress responses under pathological conditions.
INTRODUCTION
Stress granules (SGs) are ribonucleoprotein aggregates that form in response to stress conditions such as oxidative stress, osmotic stress, or heat shock (1). SGs temporarily store translationally arrested messenger RNAs that are unnecessary to survive stress conditions, shifting the translation machinery toward stress-relevant transcripts. Once SGs disassembled, the transcripts triaged from translation are released and their translation can resume.
The protein composition of SGs as well as the regulation of their formation and disassembly are only partially understood (2). The Ras GTPase-activating protein-binding protein G3BP1 (3) is a critical regulator of SG dynamics (4, 5), RNA metabolism (6–8), and translation (9, 10). It was reported that G3BP1 played a critical role in the secondary aggregation of smaller stress-induced ribonucleoprotein granules into larger SGs (5). The dynamics of G3BP1 SG formation was reported to be regulated by serine phosphorylation (4, 6, 11, 12) and arginine methylation (13).
Lysine acetylation is a major posttranslational modification impacting thousands of mammalian proteins, rivaling other major posttranslational modifications (14). However, our understanding of the roles of the acetylation of individual residues is lagging far behind that of phosphorylation. It was reported that G3BP1 and the histone deacetylase 6 (HDAC6) lysine deacetylase interacted with each other, and HDAC6 regulated SGs (15). However, the regulation of G3BP1 function by lysine acetylation has not been reported. We hypothesized that the acetylation of G3BP1 regulates SG dynamics. We carried out liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis of G3BP1 immunoprecipitated from mammalian cells and identified the lysine-376 (K376) residue as the sole acetylated lysine residue in G3BP1 in various mammalian cell lines. Our results suggest that G3BP1 acetylation is a dynamic process, since the basal level of acetylated G3BP1 was rather low in the tested cell types. The acetylation of K376 and the K376Q acetylation-mimicking mutation interfered with G3BP1 RNA binding that was necessary for efficient G3BP1 SG formation. The level of G3BP1 K376 acetylation rose concomitantly with SG disassembly, and K376-acetylated G3BP1 was detected outside disassembling SGs. We found that CBP/p300 acetylated and HDAC6 deacetylated G3BP1 at the K376 position. Taken together, our results suggest that G3BP1 K376 acetylation plays a role in SG disassembly.
RESULTS
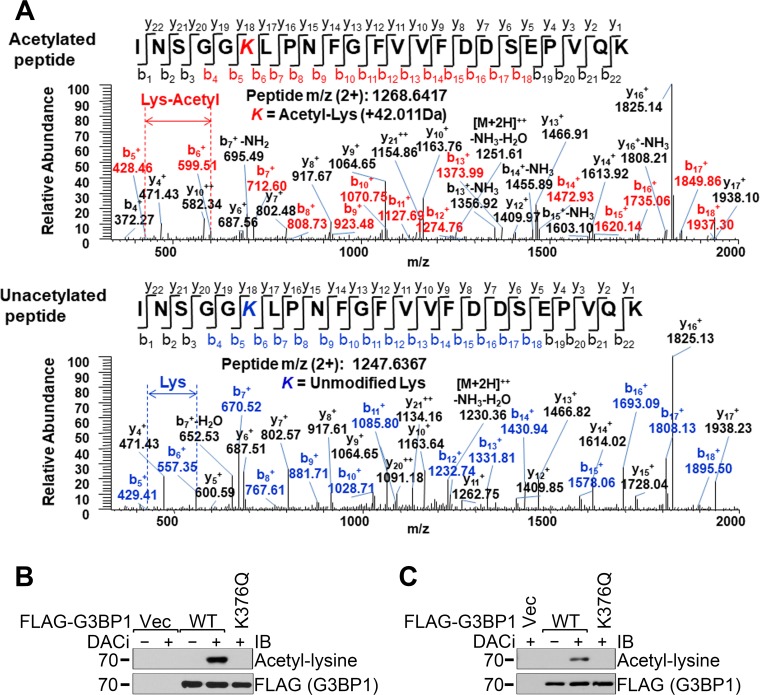
The G3BP1 protein is acetylated on the lysine-376 residue.
It was reported that G3BP1 SGs were positive for the HDAC6 lysine deacetylase, HDAC6 interacted with G3BP1, and the enzymatic activity of HDAC6 was necessary for the efficient formation of G3BP1 SGs (15). However, it has not been reported whether the G3BP1 protein itself was regulated by lysine acetylation. We carried out proteomics analysis of FLAG-G3BP1 immunoprecipitated from 293T cells treated with deacetylase inhibitors. Figure 1A unambiguously shows that lysine residue 376 (K376) was acetylated. Furthermore, replacing K376 with glutamine (K376Q) resulted in the complete loss of G3BP1 acetylation in the human embryonic kidney-derived 293T cells (Fig. 1B). It is noted that the G3BP1 acetylation level was not detectable without deacetylase inhibitors (DACi) in the cell culture medium, suggesting that G3BP1 acetylation is transient under physiological conditions. Similar results were obtained from the mouse motor neuron-like NSC-34 cells (Fig. 1C).
FIG 1.
Identification of the acetylation of the K376 residue of G3BP1. (A) Mass spectrometric identification of the acetylated and intact G3BP1 peptide INSGGK376LPNFGFVVFDDSEPVQK. (B and C) FLAG-tagged WT or K376Q G3BP1 or FLAG vector control (Vec) were transfected into 293T (B) or NSC-34 (C) cells. A day later, the cells were treated with the deacetylase inhibitors TSA, sodium butyrate, and nicotinamide as indicated overnight, followed by FLAG immunoprecipitation and immunoblotting (IB) with the indicated antibodies. Representative results of at least three independent experiments are shown.
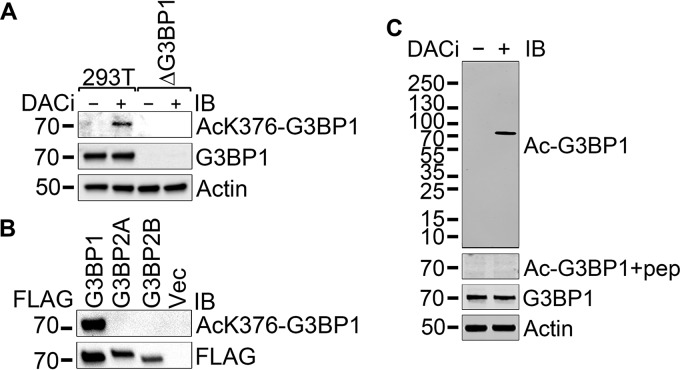
Development of an acetylated K376-G3BP1-specific antibody.
In order to investigate the role of K376 acetylation of G3BP1, we raised and affinity purified an acetylated K376-G3BP1-specific rabbit polyclonal antibody against a peptide antigen. The purified antibody detected endogenous hyperacetylated G3BP1 in 293T cells but not in G3BP1-null 293T cells in immunoblotting (Fig. 2A). The antibody detected FLAG-tagged G3BP1 immunoprecipitated from G3BP1-null 293T cells but neither isoform A nor isoform B of the closely related G3BP2 protein (Fig. 2B). The acetylated G3BP1 band became undetectable when the anti-acetylated G3BP1 antibody was blocked by preincubation with the immunizing peptide (Fig. 2C).
FIG 2.
Testing the acetylated-K376 G3BP1-specific affinity-purified rabbit polyclonal antibody. (A) 293T and G3BP1-null 293T cells were left untreated or were treated with the deacetylase inhibitors TSA, sodium butyrate, and nicotinamide overnight before lysate preparation. Immunoblotting of the extracts was performed with the indicated antibodies. (B) G3BP1-null 293T cells were transfected with FLAG-tagged G3BP1, G3BP2A, and G3BP2B expression constructs or FLAG vector control. A day later, the cells were treated with the deacetylase inhibitors TSA, sodium butyrate, and nicotinamide overnight before lysate preparation, followed by FLAG immunoprecipitation and immunoblotting with the indicated antibodies. Representative results from at least three independent experiments are shown. (C) The anti-acetylated K376 G3BP1 antibody (Ac-G3BP1) was unable to detect the acetylated G3BP1 band in DACi-treated 293T lysate when the antibody was blocked by preincubation with the immunizing peptide at a 1-μg/ml concentration for 1 h at room temperature.
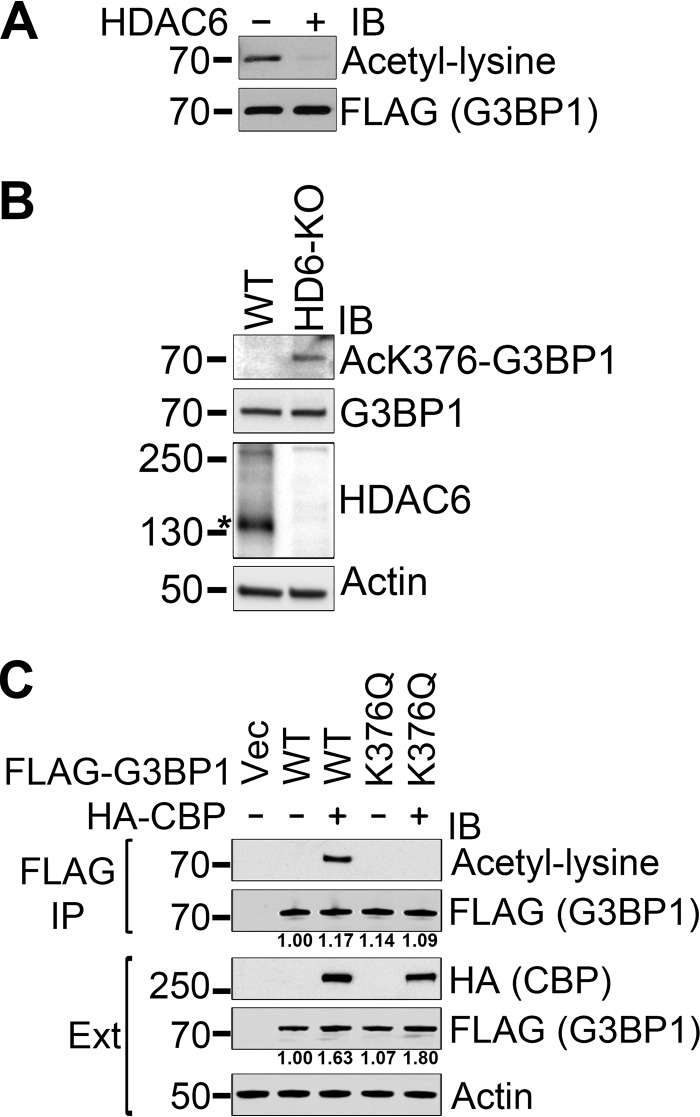
G3BP1 K376 acetylation is regulated by HDAC6 and CBP/p300.
We hypothesized that HDAC6 deacetylates G3BP1 because of their reported interaction and the localization of HDAC6 to SGs (15). We found that purified HDAC6 deacetylated G3BP1 in vitro (Fig. 3A). Immunoblotting with the anti-acetylated K376-G3BP1 antibody also detected acetylated G3BP1 in HDAC6 knockout mouse embryonic fibroblast (MEF) cells, unlike in control MEF cells (Fig. 3B).
FIG 3.
G3BP1 K376 acetylation is regulated by HDAC6 and CBP. (A) Hyperacetylated FLAG-tagged G3BP1 was immunoprecipitated from deacetylase inhibitor-treated 293T cells under native conditions and subsequently deacetylated in vitro with purified human HDAC6. (B) Endogenous G3BP1 is hyperacetylated at the K376 position in HDAC6-null MEF cells. The specific HDAC6 band is marked by an asterisk. (C) Overexpression of CBP resulted in hyperacetylation of the cotransfected FLAG-tagged WT but not K376Q mutant G3BP1. FLAG immunoprecipitation followed by immunoblotting with the indicated antibodies is shown. Representative results of at least three independent experiments are shown. The numbers under the respective panels represent the quantification of the FLAG-G3BP1 signal in the immunoprecipitations and the FLAG-G3BP1 signal normalized to the actin levels in the total extracts (Ext).
The CREB-binding protein (CBP) and its close structural and functional homolog, p300, form a unique family of protein lysine acetyltransferases that is responsible for a significant portion of protein acetylation in mammalian cells (16, 17). Whereas the acetylation of FLAG-tagged wild-type (WT) G3BP1 immunoprecipitated from 293T cells was undetectable, the overexpression of CBP resulted in clearly detectable G3BP1 acetylation that was completely abolished by the K376Q mutation (Fig. 3C). The levels of FLAG-G3BP1 in the cell extracts were higher with CBP overexpression. The WT G3BP1 level was 1.63-fold (standard deviations [SD], 6.67 × 10−2; P = 3.70 × 10−3) in the presence of CBP. The K376Q G3BP1 level was 1.69-fold (SD, 2.88 × 10−1; P = 2.83 × 10−2) in the presence of CBP. There was no statistically significant difference between WT and K376Q levels. The levels of total G3BP1 in the immunoprecipitation (IP) samples were comparable, with no statistically significant differences (Fig. 3C).
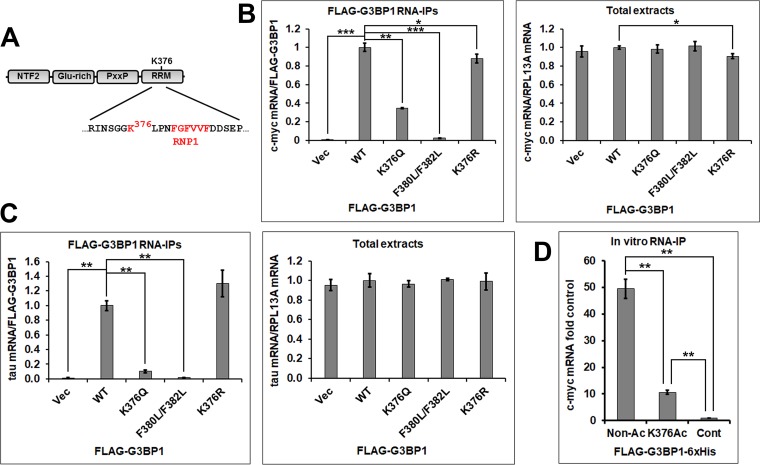
The acetylation of the K376 residue regulates G3BP1 RNA binding.
The K376 residue is located N terminally to the critical RNP1 motif (18) in the RNA-binding RRM domain of G3BP1 (Fig. 4A). Amino acid residues in positions analogous to that of K376 (loop 3 of the RRM domain) in other RRM domain-containing proteins were reported to play important roles in nucleic acid binding (18). To test whether the K376 residue and its acetylation might affect RNA binding, we performed RNA immunoprecipitation (RNA-IP) experiments with FLAG-tagged WT G3BP1, the acetylation-mimicking K376Q, and nonacetylated lysine-mimicking K376R G3BP1 mutants as bait, followed by quantitative real-time PCR quantification of the coprecipitation of two known G3BP1-binding RNA substrates, the c-myc (6) and tau (10, 19) mRNAs. The F380 and F382 residues of G3BP1 represent the conserved aromatic positions 3 and 5 of the RNP1 motif that are critical for RNA binding (18); thus, the F380L/F382L double mutant G3BP1 is RNA binding deficient (20) and was included as a negative control. When normalized to FLAG-tagged WT G3BP1, the K376Q mutant coprecipitated significantly less c-myc mRNA from G3BP1-null 293T cell extracts (34.6%; SD, 7.36 × 10−3; P = 1.23 × 10−3) (Fig. 4B). Very little c-myc mRNA was coprecipitated from cells that were transfected with the F380L/F382L double mutant (2.39%; SD, 1.57 × 10−3; P = 6.99 × 10−4) or the vector control (0.630%; SD, 7.64 × 10−4; P = 6.81 × 10−4). There were no significant differences between the c-myc mRNA levels in the total cell extracts, with the exception of the K376R mutant-transfected sample (90.6%; SD, 2.76 × 10−2; P = 1.11 × 10−2). This difference might explain why the K376R mutant G3BP1 precipitated slightly less c-myc mRNA (88.1%; SD, 4.90 × 10−2; P = 3.71 × 10−2). The WT and the K376R mutant FLAG-G3BP1 coprecipitated similar amounts of tau mRNA from NSC-34 cells, whereas significantly less tau mRNA was coprecipitated by the K376Q mutant (10.3%; SD, 1.71 × 10−2; P = 1.05 × 10−3), the F380L/F382L mutant (1.85%; SD, 1.99 × 10−3; P = 1.54 × 10−3), or the vector control (1.08%; SD, 5.58 × 10−3; P = 1.43 × 10−3) (Fig. 4C).
FIG 4.
Acetylation of the K376 residue and the K376Q acetylation-mimicking mutation impair RNA binding in RNA-IP experiments. (A) The domain structure of G3BP1. (B and C) The indicated FLAG-tagged G3BP1 expression constructs were transfected into 293T (B) or NSC-34 (C) cells. Two days later, the cells were lysed and RNA immunoprecipitation experiments were performed. The c-myc and tau mRNA amounts were normalized with the FLAG-G3BP1 levels in the FLAG-G3BP1 immunoprecipitations and with the RPL13A mRNA levels in the total extracts. Averages ± SD from three repetitions are shown. (D) In vitro RNA-IP of the c-myc mRNA from total 293T RNA with nonacetylated or K376-acetylated recombinant FLAG-G3BP1-6×His bait. Averages ± SD from three repetitions are shown. *, 0.05 > P > 0.01; **, 0.01 > P > 0.001; ***, P < 0.001.
In order to directly determine the effect of G3BP1 K376 acetylation on RNA binding, we expressed and purified recombinant FLAG-G3BP1-6×His from Escherichia coli with or without K376 acetylation using an Amber suppression-based method (21, 22). Equal amounts of the proteins were immobilized to beads and incubated with total RNA purified from 293T cells. Our results showed that K376-acetylated FLAG-G3BP1-6×His precipitated significantly less c-myc mRNA than the nonacetylated protein (21.5%; SD, 1.96 × 10−2; P = 1.80 × 10−3) (Fig. 4D). Taken together, our results show that the acetylation of the K376 residue of G3BP1 interferes with RNA binding.
The significance of K376 acetylation of G3BP1 in regulating its interactions with other SG proteins.
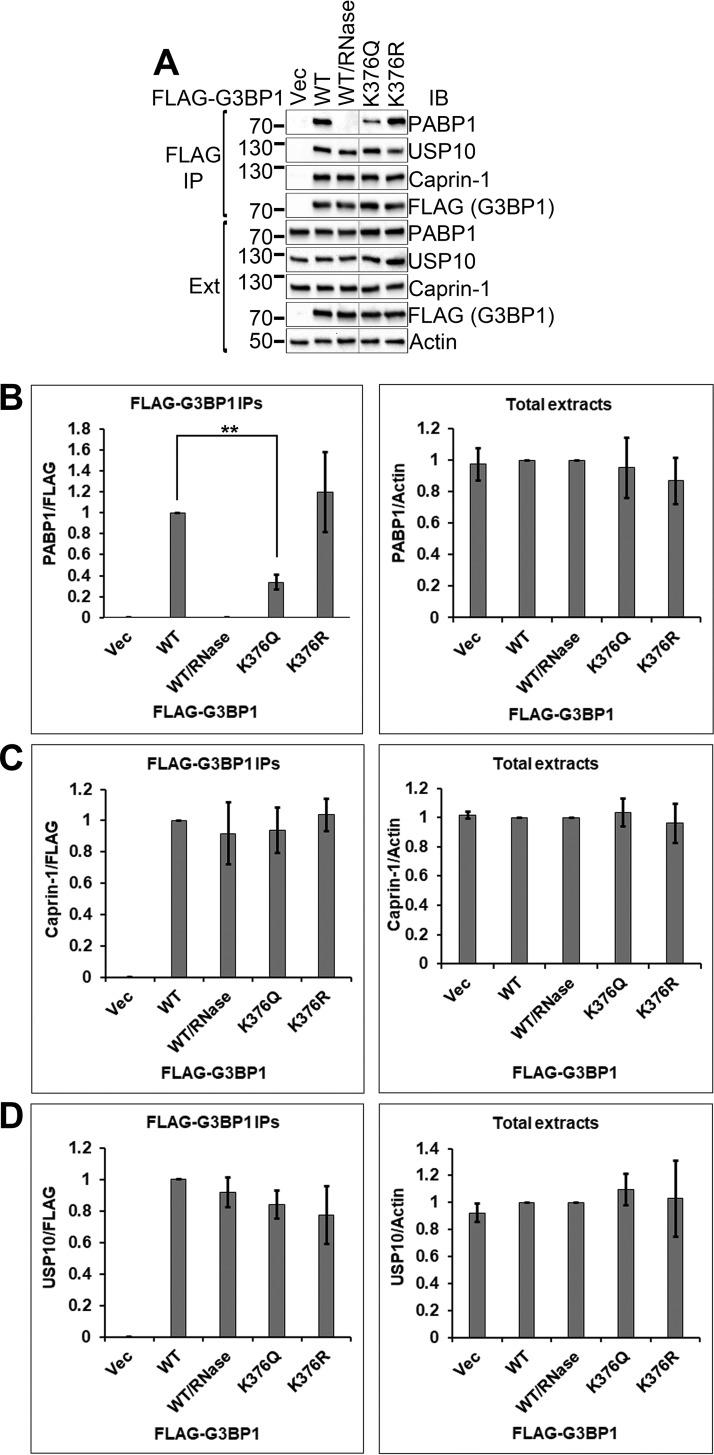
We next assessed how K376 acetylation of G3BP1 may affect its interactions with other SG proteins in coimmunoprecipitation studies. Figure 5A shows the interaction of WT or mutant G3BP1 with these proteins, and Fig. 5B to D show the quantification of the results from Fig. 5A.
FIG 5.
K376Q acetylation-mimicking mutation impaired the interaction of G3BP1 with PABP1 but not with caprin-1 or USP10. (A) The indicated FLAG-tagged G3BP1 expression constructs were transfected into 293T cells. Two days later, the cells were lysed and FLAG immunoprecipitations were performed, followed by immunoblotting with the indicated antibodies. (B to D) Quantification of experiments shown in panel A from three independent repetitions. Ext, total cell extracts. **, 0.01 > P > 0.001.
G3BP1 interacts with the SG component poly(A)-binding protein 1 (PABP1) (23). We found that the interaction between G3BP1 and PABP1 was completely abolished by the addition of RNase to the lysis buffer, consistent with previous reports that found that the interaction was RNA dependent (20, 23). The K376Q acetylation-mimicking G3BP1 mutation significantly reduced the G3BP1-PABP1 interaction (33.6%; SD, 6.95 × 10−2; P = 3.63 × 10−3). The K376R nonacetylated lysine-mimicking G3BP1 mutant interacted with PABP1 similarly to wild-type G3BP1. The total levels of the G3BP1 versions and PABP1 were comparable. Taken together, these results indicate that K376 acetylation of G3BP1 regulated the RNA-dependent G3BP1-PABP1 interaction.
The G3BP1 protein also interacts with the SG components USP10 (12, 24, 25) and caprin-1 (12, 26). We found that the G3BP1-USP10 and G3BP1–caprin-1 interactions were resistant to RNase treatment, suggesting that these interactions were independent of RNA (Fig. 5A). Neither the K376Q nor the K376R mutation had a significant effect on the interactions of G3BP1 with USP10 or caprin-1 (Fig. 5A, C, and D). Thus, K376 acetylation did not affect the RNA-independent protein-protein interactions of G3BP1, and it did not have a global impact on all interactions of G3BP1.
RNA binding and the K376 position regulate G3BP1 SG formation.
The RRM domain of G3BP1 plays a critical role in SG formation (4). It was reported that deacetylase inhibitor treatment, especially HDAC6 inhibition or HDAC6 loss, interfered with G3BP1 SG formation (15). The acetylated K376 residue is located in the RRM domain (Fig. 4A); thus, we hypothesized that G3BP1 acetylation impairs G3BP1 SG formation. To test this hypothesis, we examined SG dynamics in the presence of the WT or the acetylation-mimicking K376Q mutant G3BP1. In addition, the RNA binding-deficient F380L/F382L mutant G3BP1 was included as a control.
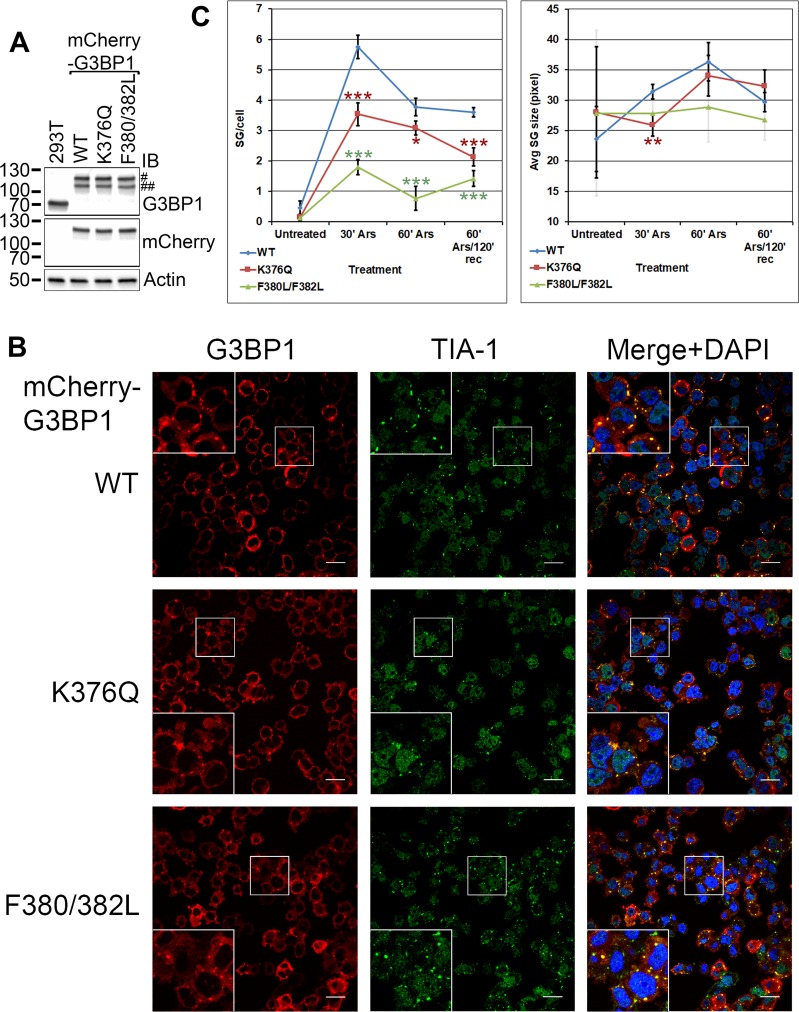
Endogenous WT G3BP1 could interfere with testing SG formation by G3BP1 mutants because of potential hetero-oligomerization. Transient transfection of G3BP1 expression constructs results in uneven expression among the cells, and cells with high expression levels spontaneously form G3BP1 granules (27). Transfection itself is an SG-inducing effect (27). To address these issues, we generated G3BP1-null 293T derivative cell lines with stably integrated mCherry-tagged WT, K376Q, or F380L/F382L G3BP1 expression constructs. We selected clones with G3BP1 expression levels close to the endogenous level (Fig. 6A) and even expression among the cells. On the G3BP1 immunoblot shown in Fig. 6A (top), the full-length mCherry-G3BP1 fusion proteins are marked by “#.” Bands at lower molecular weights (marked by “##”) were also detected in the G3BP1 immunoblot. Since only bands corresponding to the top band (#) were detected in the mCherry immunoblot, we interpret the lower bands (##) as mCherry-G3BP1 fusion proteins with partially processed mCherry tag.
FIG 6.
K376Q acetylation-mimicking mutation and the F380L/F382L RNA binding-deficient mutation impair SG formation. (A) The G3BP1-null 293T derivative mCherry-G3BP1 WT, K376Q, and F380L/F382L knock-in cell lines had G3BP1 levels comparable to that of 293T cells and each other, as shown in immunoblotting with the indicated antibodies. Symbols: #, the expected mCherry-tagged G3BP1 band; ##, mCherry-G3BP1 fusion protein with likely partially processed mCherry tag. See the text for details. (B) Representative confocal microscopic images of mCherry-G3BP1 fluorescence and TIA-1 immunofluorescence of the varied knock-in cell lines after 30 min of treatment with 0.5 mM sodium arsenite. The nuclei were visualized with DAPI stain. Scale bars, 20 μm. (C) The number ± SD of G3BP1 SGs per cell and the average size ± SD of G3BP1 SGs, as quantified in four independent experiments. The F380L/F382L error bars are shown semitransparent on the right to allow for distinction between the WT and K376Q error bars at 30 min. Significance compared to values for the WT is indicated by asterisks: *, 0.05> P > 0.01; **, 0.01 > P > 0.001; ***, P < 0.001. Rec, recovery.
Immunofluorescence imaging results showed that the G3BP1 SGs generally stained positive for another SG marker, TIA-1 (27), as well (Fig. 6B). The number of SGs per cell and the size of stress granules were quantified as shown in Fig. 6C. Very few unstressed cells harbored mCherry-G3BP1 granules that ranged widely in size (Fig. 6C). Thirty minutes after SG induction with sodium arsenite, the number of mCherry-G3BP1 granules per cell was lower with K376Q and F380L/F382L G3BP1 than with the WT (P values of 1.62 × 10−4 and 9.41 × 10−6, respectively) (Fig. 6C). By 60 min, the difference in the number of granules per cell between K376Q or F380L/F382L and WT G3BP1 remained significant (P values of 1.05 × 10−2 and 3.51 × 10−5, respectively). After 2 h of recovery in arsenite-free medium, the number of K376Q granules dropped more rapidly than that of the WT (Fig. 6C), and both the K376Q and the F380L/F382L mutants had significantly fewer granules per cell than WT G3BP1 (P values of 4.95 × 10−4 and 3.85 × 10−5, respectively). As expected, the RNA binding-deficient F380L/F382L mutation impaired SG formation more severely. Thirty minutes after induction, the sizes of the mCherry-K376Q G3BP1 granules were significantly smaller than those of mCherry-WT G3BP1 granules (P = 3.75 × 10−3), although by 60 min after induction, this difference became statistically insignificant (Fig. 6C).
Taken together, our results suggest that RNA binding was necessary for the efficient formation of G3BP1 SGs. Mimicking the acetylation of the K376 residue of G3BP1 with the K376Q mutation also interfered with G3BP1 incorporation into SGs and led to more rapid SG disassembly than that of WT G3BP1.
The acetylation of G3BP1 K376 is upregulated during SG disassembly.
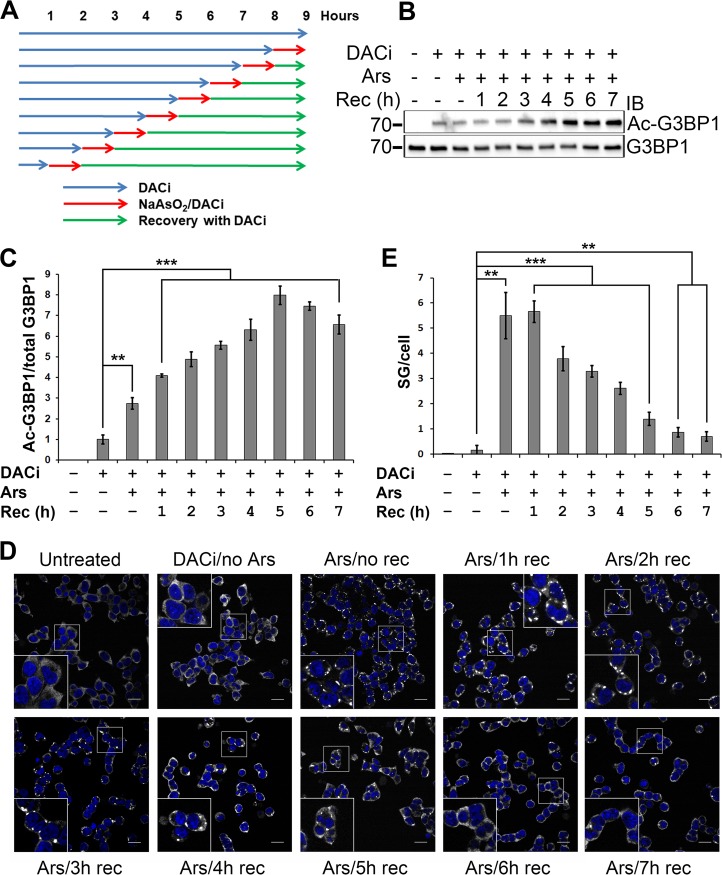
We had set forth to determine whether G3BP1 acetylation correlated with stress granule induction or disassembly. The acetylation of G3BP1 K376 was undetectable in 293T cells without deacetylase inhibitor treatment. In order to determine the time course of G3BP1 acetylation during SG induction and disassembly, 293T cells were pretreated with deacetylase inhibitors, with or without a 1-h treatment with sodium arsenite and with or without recovery in fresh medium containing deacetylase inhibitors. The total duration of the deacetylase inhibitor treatment of the cells was identical; only the start time of the 1-h sodium arsenite treatment was varied (Fig. 7A).
FIG 7.
Time course of G3BP1 acetylation during SG induction and disassembly. (A) Schematic of the treatment of 293T cells with deacetylase inhibitors and sodium arsenite (0.5 mM). The control cells were left untreated. (B) Immunoblotting of the respective cell lysates treated as described for panel A with the indicated antibodies. (C) Quantification of the specific K376 acetylation of G3BP1, normalized to the DACi-treated, sodium arsenite-untreated condition. The averages ± SD from three experiments are shown. (D) Representative confocal microscopic images of G3BP1 immunofluorescence (white pseudocoloring) at various time points as shown in panel A. The nuclei were visualized with DAPI stain. Scale bars, 20 μm. (E) Quantification of the number ± SD of G3BP1 granules per cell, as shown in panel D, from four independent experiments. **, 0.01 > P > 0.001; ***, P < 0.001. Ars, arsenite; Rec, recovery.
Sodium arsenite treatment without recovery significantly elevated specific G3BP1 acetylation (the K376-acetylated G3BP1/total G3BP1 ratio) compared to no arsenite treatment (P = 1.37 × 10−3) (Fig. 7B and C). Allowing the cells to recover in fresh medium after arsenite treatment resulted in further elevation of G3BP1 K376 acetylation peaking at 5 h of recovery, after which specific acetylation declined (Fig. 7B and C), probably due to a combination of de novo G3BP1 synthesis and the turnover of acetylated G3BP1.
In order to determine how G3BP1 acetylation correlated with the dynamics of G3BP1 SGs, we performed G3BP1 immunofluorescence experiments on cells treated as described for Fig. 7A. It was found that the increase in G3BP1 acetylation correlated with the disassembly of G3BP1 granules (Fig. 7D and E).
The regulation of the interactions of G3BP1 with other SG proteins under stress conditions.
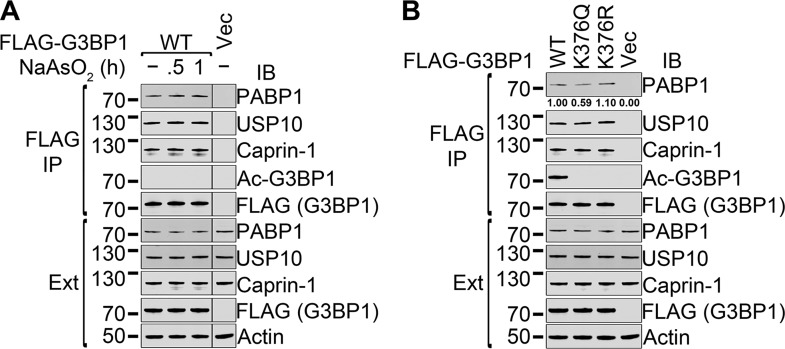
To determine whether the interactions of G3BP1 with other SG proteins change under stress conditions, FLAG-tagged WT G3BP1 was transfected into 293T cells. Two days later, the cells were either left untreated or were treated for 30 min or 1 h with 0.5 mM sodium arsenite in the medium, followed by FLAG immunoprecipitation. We did not find any statistically significant difference in the coprecipitation of PABP1, USP10, or caprin-1 with FLAG-G3BP1 in three repetitions (Fig. 8A). Without deacetylase inhibitors, the acetylation of G3BP1 was not detectable. The results showed that the interaction between G3BP1 and stress granule proteins was not changed under stress.
FIG 8.
Regulation of the interactions of G3BP1 with other SG proteins under stress conditions. (A) The FLAG-tagged WT G3BP1 expression construct or vector control was transfected into 293T cells. Two days later, the cells were either left untreated or were treated for 30 min or 1 h with 0.5 mM sodium arsenite in the medium, followed by FLAG immunoprecipitation and immunoblotting with the indicated antibodies. Representative images of three repetitions are shown. (B) The FLAG-tagged WT, K376Q, or K376R G3BP1 expression construct or vector control was transfected into 293T cells. Two days later, the cells expressing FLAG-G3BP1 were treated for 1 h with 0.5 mM sodium arsenite in the medium, followed by 5 h of recovery without sodium arsenite in the presence of deacetylase inhibitors (DACi). The cells were lysed and FLAG immunoprecipitations were performed, followed by immunoblotting with the indicated antibodies. Under the top panel, the relative quantification of the coprecipitated PABP1 protein is shown from three repetitions. Only the coprecipitation of PABP1 with WT or K376Q G3BP1 was statistically significantly different (see the text for details). Ext, total cell extracts.
We next performed immunoprecipitations with the WT and K376Q and K376R FLAG-G3BP1 mutants following treatment with 0.5 mM sodium arsenite for 1 h and 5 h of recovery in arsenite-free medium in the presence of deacetylase inhibitors (Fig. 8B). The reason to choose this condition was that based on the results shown in Fig. 7, the level of G3BP1 acetylation reached a maximum 5 h posttreatment with arsenite. Comparable levels of coprecipitated caprin-1 and USP10 were observed with all FLAG-G3BP1 baits. The amounts of PABP1 coprecipitated with WT G3BP1 and the K376R mutant also were not statistically significantly different. A smaller amount of PABP1 was coprecipitated with K376Q G3BP1 than with the WT (58.9%; SD, 6.02 × 10−2; P = 7.09 × 10−3). This difference was smaller than that seen with unstressed cells (33.6%; SD, 6.95 × 10−2; P = 3.63 × 10−3) (Fig. 5), consistent with the observation that G3BP1 K376 acetylation decreased the RNA-dependent interaction between G3BP1 and PABP1.
The inhibition of G3BP1 acetylation delays stress granule disassembly.
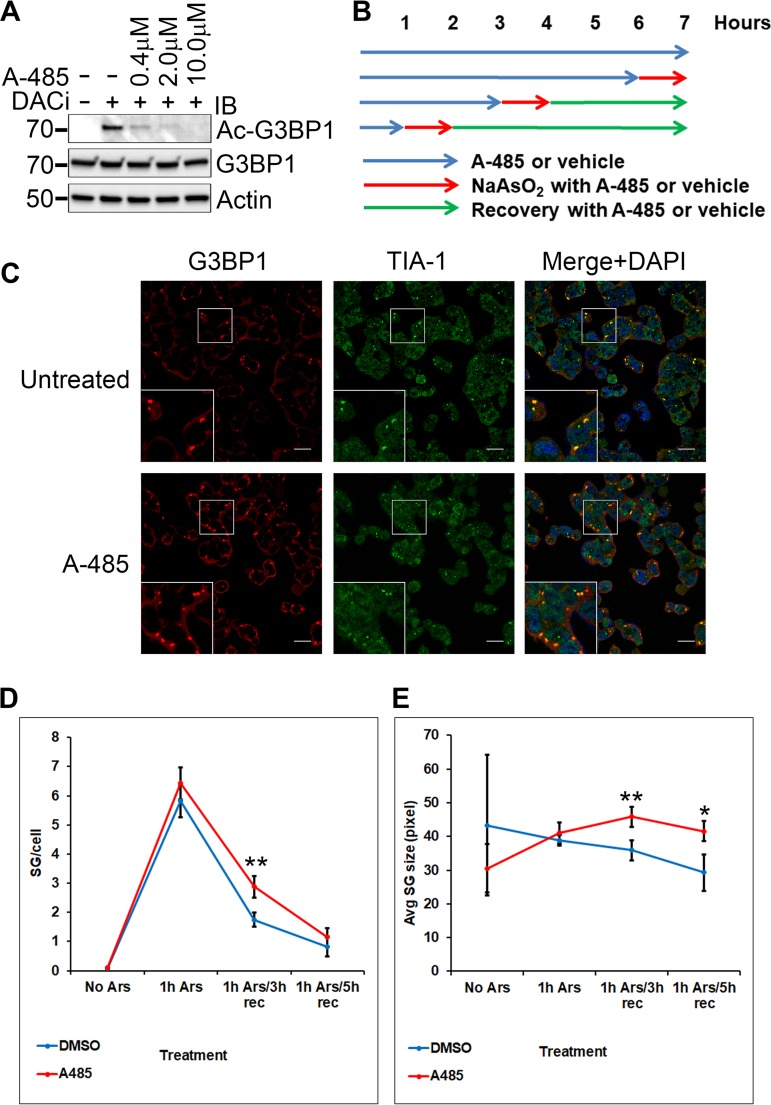
To determine whether endogenous CBP/p300 played a role in G3BP1 acetylation upon SG induction and disassembly, 293T cells were treated with sodium arsenite with or without pretreatment with deacetylase inhibitors and the highly selective CBP/p300 inhibitor A-485 (28), followed by 5 h of recovery in the respective arsenite-free media (Fig. 9A). We found that A-485 significantly inhibited G3BP1 K376 acetylation (Fig. 9A). Our results show that endogenous CBP/p300 plays a major role in G3BP1 K376 acetylation.
FIG 9.
Effect of CBP/p300 inhibition on G3BP1 stress granule dynamics. (A) 293T cells were pretreated with deacetylase inhibitors and the indicated concentrations of the highly selective CBP/p300 inhibitor A-485 (6387; Tocris) as shown for 1 h in the medium, followed by 1 h of SG induction with 0.5 mM sodium arsenite and 5 h of recovery in the respective fresh medium with DACi and A-485. The control cells were treated with sodium arsenite only. The cell lysates were subjected to immunoblotting with the indicated antibodies. (B) Schematic of the treatment of 293T cells with A-485 (10 μM) or vehicle (DMSO) and sodium arsenite (0.5 mM). (C) Representative confocal microscopic images of G3BP1 and TIA-1 immunofluorescence of 293T cells treated with sodium arsenite for 1 h, followed by recovery in the respective fresh media for 3 h. The nuclei were visualized with DAPI stain. Scale bars, 20 μm. (D and E) The number ± SD of G3BP1 SGs per cell (D) and the average size ± SD of G3BP1 SGs (E), as quantified in four independent experiments. *, 0.05 > P > 0.01; **, 0.01 > P > 0.001.
In order to determine whether the inhibition of acetylation affected G3BP1 stress granule dynamics, 293T cells were treated with sodium arsenite in the presence of A-485 or vehicle (dimethyl sulfoxide [DMSO]), either without or with 3 or 5 h of recovery in the respective fresh media (Fig. 9B shows a schematic of this method). Without recovery, neither the average number of G3BP1 SGs per cell nor their average size was different with or without A-485 treatment. However, when stress granule induction was followed by 3 h of recovery, the number of stress granules per cell was significantly higher in the presence of A-485 (P = 3.11 × 10−3) (Fig. 9C and D). By 5 h of recovery, the difference between the number of stress granules per cell became insignificant (Fig. 9D). The average size of the G3BP1 granules was significantly larger in the presence of A-485 at both 3 and 5 h of recovery (P values of 3.24 × 10−3 and 1.25 × 10−2, respectively) (Fig. 9E). Taking these findings together, the inhibition of acetylation by A-485 delayed the disassembly of G3BP1 stress granules.
Acetylated G3BP1 is localized primarily outside SGs.
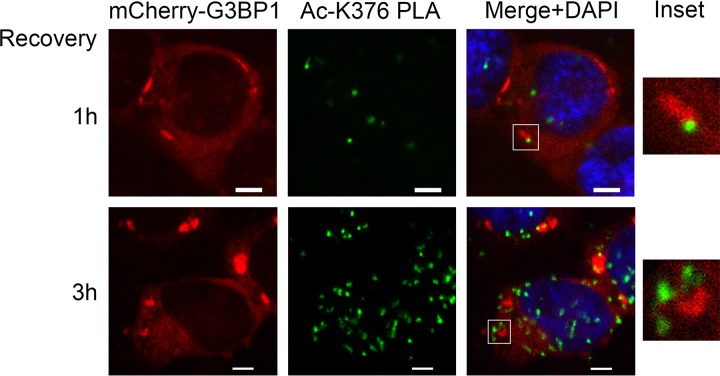
To ensure the specificity of the anti-acetylated K376-G3BP1 signal, we performed proximity ligation assay (PLA) (29) using a mouse anti-G3BP1 antibody and the rabbit anti-K376-acetylated G3BP1 antibody. In PLA, signal is only generated when the distance between the two antibodies used is not more than 40 nm. 293T cells stably expressing mCherry-tagged WT G3BP1 were pretreated with deacetylase inhibitors for 1 h. Stress granules were induced with sodium arsenite for 1 h in the presence of DACi, followed by 1 or 3 h of recovery in fresh medium containing DACi. The acetylated G3BP1 foci were localized mostly outside the G3BP1 SGs, often on their periphery (Fig. 10). As a negative control, either one or the other antibody was omitted, resulting in complete loss of the PLA signal.
FIG 10.
K376-acetylated G3BP1 localizes primarily outside SGs. Representative confocal microscopic images of mCherry-G3BP1 fluorescence and the K376-acetylated G3BP1 PLA foci (Ac-K376 PLA). The nuclei were visualized with DAPI stain. Acetylated G3BP1/total G3BP1 proximity ligation assay was performed on G3BP1-null 293T-derived cells stably expressing mCherry-WT G3BP1. The cells were pretreated with deacetylase inhibitors for 1 h, followed by SG induction with 0.5 mM sodium arsenite for 1 h and 1 or 3 h of recovery in fresh medium containing deacetylase inhibitors. The acetylated G3BP1 PLA foci were often found in the periphery of the disassembling SGs. The insets show magnifications of the boxed areas. Scale bars, 5 μm.
DISCUSSION
The G3BP1 protein is a critical regulator of SG dynamics (4, 5), an important adaptive process whose regulation is not fully understood. It was reported that G3BP1 interacted with the major cytoplasmic lysine deacetylase HDAC6 and that HDAC6 inhibition or loss interfered with G3BP1 SG formation (15). We hypothesized that the function of G3BP1 was regulated through its acetylation, controlled by HDAC6. First, we identified the K376 residue as the single G3BP1 lysine acetylation site in the 293T cell line and in the developing mouse motor neuron-like NSC-34 cells (Fig. 1). We also detected K376-acetylated G3BP1 in mouse embryonic fibroblast cells (Fig. 3B). This finding is supported by other proteomic studies that also identified K376 acetylation of G3BP1 in various cells (30–32).
The acetylated K376 residue of G3BP1 is in the RNA binding RRM domain, immediately N terminal to the conserved FGFVVF motif that is critical for RNA binding (Fig. 4A). Thus, we hypothesized that K376 acetylation plays a role in the regulation of RNA binding. Indeed, we found that the K376Q acetylation-mimicking mutation and K376 acetylation decreased RNA binding in RNA-IP experiments (Fig. 4B to D). The RNA-dependent interaction of G3BP1 with PABP1 was also impacted by the K376Q mutation, whereas the RNA-independent protein-protein interactions of G3BP1 with USP10 and caprin-1 were little affected (Fig. 5).
The RRM domain of G3BP1 plays a critical role in SG formation (4). We reported that the F380L/F382L double mutation in the RRM domain impaired G3BP1 RNA binding (20), and here we report that it also severely inhibited SG formation (Fig. 6). Consistent with the role of the K376 residue in RNA binding, the acetylation-mimicking K376Q mutation interfered with SG formation and also led to faster SG disassembly than that of WT G3BP1 (Fig. 6).
The acetylation of the K376 residue of G3BP1 is a dynamic process, since in our experiments it was undetectable without deacetylase inhibitor treatment. It was reported that G3BP1 SGs were HDAC6 positive, HDAC6 interacted with G3BP1, and the enzymatic activity of HDAC6 was necessary for the efficient formation of G3BP1 SGs (15). We found that HDAC6 deacetylated the G3BP1 protein, and G3BP1 was hyperacetylated in HDAC6 knockout MEF cells (Fig. 3A and B). Our findings are supported by a lysine acetylation proteomics study (31) that found that G3BP1 K376 acetylation was primarily enhanced by pan-HDAC inhibitors and the selective HDAC6 inhibitor tubacin (33).
The CBP protein and its close homolog, p300, are major protein lysine acetyltransferases (16, 17). Overexpressed CBP specifically acetylated the K376 residue of G3BP1 (Fig. 3C). The highly selective CBP/p300 inhibitor A-485 (28) abrogated G3BP1 acetylation (Fig. 9A), showing that endogenous CBP/p300 was the major G3BP1 acetyltransferase.
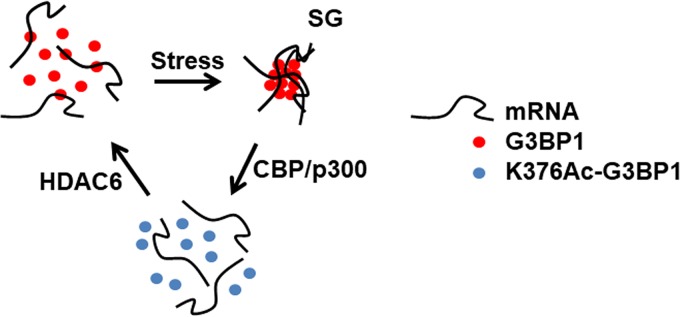
Our results show that the induction of SGs facilitated G3BP1 acetylation (Fig. 7). However, the highest level of acetylation was detected in the recovery phase after SG induction, correlating with SG disassembly (Fig. 7). The inhibition of acetylation resulted in delayed disassembly of G3BP1 stress granules (Fig. 9B to E). The acetylation of G3BP1 K376 interfered with RNA binding (Fig. 4 and 5) and SG formation (Fig. 6). The K376-acetylated G3BP1 foci localized primarily outside SGs (Fig. 10). Based on the above-described results, we propose a model regarding the role of G3BP1 K376 acetylation in the regulation of SG dynamics (Fig. 11). Under unstressed conditions, G3BP1 and its bound mRNA substrates are dispersed throughout the cytoplasm. Upon stress, the G3BP1-containing ribonucleoproteins are concentrated into SGs. The acetylation of G3BP1 by CBP/p300 results in decreased RNA binding, facilitating the disassembly of SGs. The regeneration of G3BP1 into its RNA binding state is mediated by HDAC6. Our model suggests that HDAC6 deletion or inhibition would result in G3BP1 hyperacetylation and, in turn, impairment of SG induction. This could at least partly explain the impairment of SG induction by HDAC6 inhibition in a previous study (15).
FIG 11.
Proposed model of the regulation of stress granule dynamics through G3BP1 K376 acetylation by HDAC6 and CBP/p300.
Our results with the K376Q G3BP1 mutant support our conclusions about the role of G3BP1 acetylation in stress granule dynamics (Fig. 6). However, stress granules contain a variety of other proteins as well (2). It is conceivable that other SG components could also be modified by lysine acetylation, impacting the dynamics of stress granules.
The normal dynamics of SGs is critical for the survival of stressed neuronal cells (5). Altered SG dynamics, especially impaired disassembly of SGs and the consequent formation of persistent SG-like structures, are particularly detrimental to neurons. Permanent stress granule-like structures were implicated in a range of neurodegenerative diseases, including amyotrophic lateral sclerosis (5, 20, 34–39), Parkinson’s disease (40), and Alzheimer’s disease (41). It was recently reported that the disassembly of axonal SG-like G3BP1 structures accelerated nerve regeneration in vivo (42). The HDAC6 lysine deacetylase was implicated in neurodegeneration (43–45), and the inhibition of HDAC6 was reported to be beneficial in neurodegenerative diseases (43, 46–52). Our results suggest that G3BP1 hyperacetylation and the inhibition of SG formation contribute to the beneficial effect of HDAC6 inhibition countering neurodegeneration. G3BP1 is overexpressed in several human cancers and facilitates cellular proliferation, mediated in part by its association with RNA (53). The development of targeted strategies to enhance G3BP1 K376 acetylation could provide therapeutic avenues in diseases in which hyperactive SG formation or G3BP1 overexpression is a component of the pathogenesis.
MATERIALS AND METHODS
Plasmids.
The FLAG-G3BP1, FLAG-G3BP2A, and FLAG-G3BP2B plasmids (54) were generous gifts from Zhi-Min Yuan (University of Texas Health Science Center at San Antonio). The K376Q, K376R, and K376(amber stop) mutations were introduced with the QuikChange II site-directed mutagenesis kit (Agilent). The FLAG-G3BP1, F380L/F382L construct was reported before (20). The mCherry-tagged wild type and the K376Q and F380L/F382L mutant G3BP1 constructs were made by subcloning the respective G3BP1 fragments into pmCherry-C1 (Clontech). To create mCherry-G3BP1 knock-ins, the mCherry-G3BP1 fragments were subcloned into pcDNA3.1/Hygro-Δ58, a derivative of pcDNA3.1/Hygro(+) (Thermo Fisher Scientific) in which the simian virus 40 (SV40) origin of replication was inactivated by a 58-nucleotide deletion without inactivating the SV40 early promoter (corresponding to deletion Δ58 [55]). The CBP-HA plasmid was a generous gift from Chunming Liu (College of Medicine, University of Kentucky) (56, 57). The pBK-AcKRS3 and pCDF-PylT plasmids (21, 22) used to generate recombinant nonacetylated and K376-acetylated G3BP1 were generous gifts from Jason W. Chin (Medical Research Council, Laboratory of Molecular Biology, England, UK). The wild-type and the K376(amber stop) G3BP1 coding sequences were subcloned into pCDF-PylT, arranged as N-FLAG(DYKDDDDK)-G3BP1-6×His-C. All plasmid constructs were verified with sequencing.
Cell culture and transfection.
The spinal cord neuron × neuroblastoma hybrid cell line NSC-34 (generous gift from Neil R. Cashman, University of British Columbia, Vancouver, Canada) (58, 59) and HEK293T cells (293T; ATCC CRL-3216) (60) were cultured in high-glucose Dulbecco’s modified Eagle’s medium (DMEM; D5796; Millipore Sigma) supplemented with 10% fetal bovine serum (FBS) and penicillin-streptomycin at 37°C in 5% CO2, 95% air with humidification. The NSC-34 and 293T cells were transfected with Lipofectamine 2000 (11668; Thermo Fisher Scientific) and PEI MAX (24765; Polysciences, Inc.), respectively. The wild-type and HDAC6 knockout (KO) mouse embryonic fibroblast cells (61) (generous gifts from Tso-Pang Yao, Duke University School of Medicine) were cultured in high-glucose DMEM containing sodium pyruvate (D6429; Millipore Sigma), 10% FBS, and penicillin-streptomycin at 37°C in 5% CO2, 95% air with humidification.
IP.
Cell lysates were prepared in 1× radioimmunoprecipitation assay (RIPA) lysis buffer (20-188; Millipore Sigma) supplemented with protease inhibitor cocktail (P8340, 1:500; Millipore Sigma), sodium orthovanadate (1 mM), nicotinamide (20 mM), sodium butyrate (20 mM), and trichostatin A (TSA) (1.5 μM). The cell lysates were homogenized by passing through a 23-gauge needle several times. Immunoprecipitations (IP) were performed at 4°C for 2 h using 4 μg/ml mouse anti-FLAG M2 antibody (F3165; Millipore Sigma) and Protein G UltraLink resin (53126; Thermo Fisher Scientific). The beads were washed three times with lysis buffer. The bound proteins were eluted with 0.15 μg/μl 3× FLAG peptide (F4799; Millipore Sigma) in 1× RIPA buffer at 4°C for 1 h.
Mass spectrometric identification of G3BP1 K376 acetylation.
FLAG-tagged G3BP1 was transfected into 293T cells. One day posttransfection, the transfected cells were treated with a cocktail of lysine deacetylase inhibitors containing nicotinamide (30 mM), sodium butyrate (50 mM), and trichostatin A (3 μM) overnight in regular medium, followed by anti-FLAG immunoprecipitation the following day. The eluates were resolved by denaturing polyacrylamide gel electrophoresis (SDS-PAGE), followed by staining with SYPRO Ruby protein gel stain (S12000; Thermo Fisher Scientific). The band corresponding to FLAG-G3BP1 (electrophoretic mobility of approximately 70 kDa) was excised, followed by dithiothreitol reduction, iodoacetamide alkylation, and in-gel trypsin digestion, and was analyzed as published previously (62). Briefly, the resulting tryptic peptides were extracted, concentrated, and subjected to liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis using an LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific) coupled with a Nanoflex cHiPLC system (Eksigent) through a nano-electrospray ionization source. The peptide samples were separated with a reversed-phase cHiPLC column using a gradient of water with 0.1% (vol/vol) formic acid and acetonitrile with 0.1% (vol/vol) formic acid. The LC-MS/MS data were submitted for MS/MS protein identification using the MASCOT algorithm via Proteome Discoverer (version 1.3; Thermo Fisher Scientific), applying a custom database containing human protein sequences from UniProt (63). The MASCOT MS/MS ion search was performed allowing a maximum of two missed tryptic cleavages. The considered amino acid modifications included lysine acetylation, cysteine carbamidomethylation, methionine oxidation, as well as serine, threonine, and tyrosine phosphorylation. The mass error tolerance was set to be less than 10 ppm for MS and 0.8 Da for MS/MS. A decoy database was built and searched to determine the false discovery rates (FDR). Peptides with FDR lower than 0.01 were assigned as high-confidence identifications. The mass spectra reported in this study were acquired at the University of Kentucky Proteomics Core Facility (https://www.research.uky.edu/proteomics-core-facility).
Generation of the anti-K376-acetylated G3BP1 antibody.
The nonacetylated N-CRINSGGK376LPNFG-C and acetylated N-CRINSGG(K376ac)LPNFG-C G3BP1 peptides were synthesized with C-terminal amidation (GenScript, Piscataway, NJ), with the inclusion of an extra N-terminal cysteine residue to facilitate conjugation to keyhole limpet hemocyanin (KLH) and the SulfoLink immobilization kit for peptides (44999; Thermo Fisher Scientific). The rabbit polyclonal anti-K376-acetylated G3BP1 antibody was generated by immunizing rabbits with the acetylated peptide antigen conjugated to KLH (Pocono Rabbit Farm and Laboratory, Canadensis, PA). The serum was first depleted with SulfoLink-conjugated nonacetylated peptide, and then the acetylated G3BP1 antibodies were purified with SulfoLink-conjugated acetylated peptide. The antibodies were eluted with 0.1 M glycine-HCl buffer, pH 2.5, neutralized by the addition of 1 M Tris-HCl, pH 8.5, and dialyzed against storage buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 10% glycerol, 0.04% [wt/vol] sodium azide).
Generation of nonacetylated and K376-acetylated recombinant G3BP1.
pBK-AcKRS3 and the wild-type or K376(Amber) pCDF-PylT-FLAG-G3BP1-6×His plasmids were transformed into Rosetta2(DE3)pLysS Escherichia coli cells (71-401-4; Millipore). The transformed cells were inoculated into LB medium supplemented with spectinomycin (50 μg/ml) and kanamycin (50 μg/ml) and grown with vigorous shaking at 37°C until mid-log phase. The cultures were supplemented with 20 mM nicotinamide and the K376(Amber) culture along with 10 mM N-epsilon-acetyl-l-lysine (A7231; ApexBio) and shaken for a further 30 min before induction with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). The cultures were shaken for 3 more hours, harvested by centrifugation, washed with 1× phosphate-buffered saline (PBS) containing 20 mM nicotinamide, and frozen at −80°C. The cell lysates were prepared by resuspending the pellets in 5 ml lysis buffer (50 mM NaH2PO4, 50 mM NaCl, 10 mM imidazole [pH 8.0], 1 tablet/10 ml cOmplete Mini EDTA-free protease inhibitor cocktail [11836170001; Roche], phenylmethanesulfonyl fluoride [0.2 mM], nicotinamide [20 mM], and DNase I [5 μg/ml]) per gram of wet weight. The lysates were incubated on ice until the viscosity decreased. The lysates were cleared by centrifugation at 16,000 × g at 4°C for 30 min. The FLAG-G3BP1-6×His proteins were purified from the supernatant with nickel-nitrilotriacetic acid–agarose (30210; Qiagen) by following the manufacturer’s instructions.
RNA-IP.
The RNA-IPs were performed similarly to immunoprecipitations with the inclusion of SUPERase-In RNase inhibitor (AM2696; 0.2 U/μl; Thermo Fisher Scientific) in the cell lysates and the elution buffer. Total RNA was extracted from aliquots of the cellular extracts and RNA-IP eluates with TRIzol LS reagent (10296; Thermo Fisher Scientific). To perform in vitro RNA-IP with recombinant unmodified or K376-acetylated FLAG-G3BP1-6×His, 0.6 μg of each protein was immobilized to 30 μl packed Protein G UltraLink resin using 5 μg mouse anti-FLAG M2 antibody in 1× RIPA buffer containing 0.2 μg/μl bovine serum albumin (BSA; B9000S; molecular biology grade; New England Biolabs). In the control experiments, no recombinant G3BP1 was added. The beads were washed and suspended in 300 μl micrococcal nuclease (MN) buffer (50 mM Tris-HCl, 5 mM CaCl2, pH 7.9) and incubated with 2,000 gel units of MN (M0247S; New England Biolabs) at 37°C with mild agitation to degrade potential E. coli-derived nucleic acids. The MN was inactivated with the addition of EGTA in a 2-fold molar excess over CaCl2, followed by 10 min of incubation at room temperature. The beads were washed with 1× RIPA buffer. The beads were suspended in 400 μl 1× RIPA buffer containing 0.2 μg/μl molecular biology-grade BSA, SUPERase-In RNase inhibitor (0.4 U/μl), 1 μg/μl tRNA (R5636; Millipore Sigma), and 12.5 ng/μl total RNA isolated from 293T cells using TRIzol reagent (15596; Thermo Fisher Scientific), and then they were incubated at room temperature for 1 h with mild end-over-end shaking. The beads were washed three times with 1× RIPA buffer, and the bound proteins were eluted with 3×FLAG peptide in 1× RIPA buffer. The bound RNA was extracted with TRIzol LS reagent from the eluates.
The RNAs were reverse transcribed with the SuperScript III first-strand synthesis system (18080; Thermo Fisher Scientific) using the random hexamer method and subjected to quantitative real-time PCR using Power SYBR green PCR master mix (4367659; Thermo Fisher Scientific). The quantitative PCR primers and annealing temperatures were the following: human c-myc, 5′-CACAAACTTGAACAGCTACGG-3′ and 5′-GGTGATTGCTCAGGACATTTC-3′ (9), 55°C; human RPL13A, 5′-GCCATCGTGGCTAAACAGGTA-3′ and 5′-GTTGGTGTTCATCCGCTTGC-3′ (64), 55°C; mouse tau, 5′-GGTCGAAGATTGGCTCTACTG-3′ and 5′-GCCAAGGAAGCAGACACTTC-3′ (19), 57°C; and mouse RPL13A, 5′-CTGTGAAGGCATCAACATTTCTG-3′ and 5′-GACCACCATCCGCTTTTTCTT-3′ (65), 55°C.
Immunoblotting.
Proteins were resolved by SDS-PAGE and transferred to nitrocellulose membranes. The membranes were blocked and antibodies were applied in 5% nonfat dry milk in TBST (100 mM Tris-HCl [pH 7.5], 0.9% NaCl, 0.1% Tween 20), with the exception of the rabbit anti-acetylated lysine antibody that was applied in 5% bovine serum albumin (BSA) in 20 mM Tris-HCl [pH 7.5], 0.8% NaCl, 0.1% Tween 20. The anti-acetylated lysine and the anti-K376-acetylated G3BP1 antibodies were applied in the presence of deacetylase inhibitors (20 mM nicotinamide, 1.5 μM trichostatin A). The antibodies used were rabbit anti-G3BP1 (13057-2-AP; Proteintech), mouse anti-FLAG M2-HRP (A8592; Millipore Sigma), rabbit anti-acetylated-lysine (9441; Cell Signaling Technology), rabbit anti-PABP1 (ab21060; Abcam), rabbit anti-caprin-1 (15112-1-AP; Proteintech), rabbit anti-USP10 (5553S; Cell Signaling Technology), rabbit anti-mCherry (ab167453; Abcam), mouse anti-β-actin (sc-81178; Santa Cruz Biotechnology), and the custom-made rabbit anti-K376-acetylated G3BP1 antibody.
Generation of the mCherry-G3BP1 knock-in cell lines.
The generation of the G3BP1-null 293T cells was reported before (20). The pcDNA3.1/Hygro-Δ58-mCherry-G3BP1 wild-type, K376Q, and F380L/F382L plasmids were linearized with PciI restriction endonuclease and transfected into G3BP1-null 293T cells. Cell lines with stable integration of mCherry-G3BP1 were selected in the presence of 200 μg/ml hygromycin and monitored by mCherry fluorescence. Individual clones were isolated by serial dilution, and the G3BP1 expression levels of the clones were compared to each other and to that of endogenous G3BP1 of 293T cells with immunoblotting using anti-mCherry and anti-G3BP1 antibodies.
Immunofluorescence, fluorescence microscopy, and image analysis.
The cells were seeded on gelatin-treated glass coverslips. The cells were treated with 0.5 mM sodium arsenite and/or deacetylase inhibitors in fresh medium as indicated at 37°C, with or without recovery in fresh medium for the indicated times. The cells were rinsed with 1× PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4), fixed with 4% formaldehyde in 1× PBS, and permeabilized with 1× PBS supplemented with 0.25% (vol/vol) Triton X-100. The coverslips were blocked with 10% (wt/vol) BSA in 1× PBS, and the antibodies were applied in 3% (wt/vol) BSA in 1× PBS. In the immunofluorescence experiments the primary antibodies were rabbit anti-G3BP1 (13057-2-AP; Proteintech), goat anti-TIA1 (sc-1751; Santa Cruz Biotechnology) (Fig. 6B), and mouse anti-TIA1 (sc-166247; Santa Cruz Biotechnology) (Fig. 9C). The secondary antibodies were Alexa Fluor 568-conjugated donkey anti-rabbit (A10042; Thermo Fisher Scientific), Alexa Fluor 488-conjugated donkey anti-goat (A11055; Thermo Fisher Scientific), and Alexa Fluor 488-conjugated donkey anti-mouse (A21202; Thermo Fisher Scientific) antibodies. The samples were treated with 4′,6-diamidino-2-phenylindole (DAPI), mounted with Vectashield mounting medium (H-1000-10; Vector Laboratories), and visualized using a Nikon A1 confocal microscope with a 60× objective. Z-stack images of random view fields were acquired with identical imaging parameters. Maximum intensity projections of the Z stacks were analyzed for G3BP1 SG formation as published previously (20, 66) using ImageJ (http://imagej.nih.gov/ij).
Proximity ligation assay.
The G3BP1-null 293T-derived cell line expressing stably integrated mCherry wild-type G3BP1 was seeded on gelatin-treated glass coverslips. The cells were pretreated with the deacetylase inhibitors nicotinamide (30 mM), sodium butyrate (50 mM), and trichostatin A (3 μM) for 2 h in fresh medium, followed by SG induction by the addition of 0.5 mM sodium arsenite for 1 h and recovery in deacetylase inhibitor-containing fresh medium for the indicated times. Proximity ligation assay was performed using Duolink in situ PLA probes anti-rabbit plus, anti-mouse minus, and green detection reagent (DUO92002, DUO92004, and DUO92014, respectively; Millipore Sigma) by following the manufacturer’s protocol. The antibodies used were mouse anti-G3BP1 (05-1938; Millipore Sigma) and rabbit anti-K376-acetylated G3BP1 at 1-μg/ml final concentration. As a negative control, either one or the other antibody was omitted, resulting in complete loss of the PLA signal. The images were acquired using a Nikon A1 confocal microscope with a 60× objective.
ELISA quantification of G3BP1 acetylation.
293T cells at approximately 90% confluence were treated with the deacetylase inhibitors nicotinamide (30 mM), sodium butyrate (50 mM), and trichostatin A (3 μM) for 9 h in fresh medium with or without a 1-h treatment with 0.5 mM NaAsO2 in the presence of deacetylase inhibitors, as shown in Fig. 7A. The control cells were treated with fresh medium only, without deacetylase inhibitors. Cellular extracts were prepared in 1× RIPA buffer (20-188; Millipore Sigma) supplemented with protease inhibitor cocktail (P-8340; 1:500; Millipore Sigma), 1 mM sodium orthovanadate, nicotinamide (20 mM), sodium butyrate (20 mM), and trichostatin A (1.5 μM). The capture antibody, mouse anti-G3BP1 (05-1938; Millipore), was immobilized to a microplate (3590; Corning-Costar), and the plate was blocked with 5% milk in 1× PBS. The cellular extracts and a serial dilution of recombinant K376-acetylated G3BP1 were added to the wells and incubated at room temperature. The detection antibody, rabbit anti-K376-acetylated G3BP1, and the secondary horseradish peroxidase-conjugated anti-rabbit antibody (NA934; GE Life Sciences) were applied in 5% milk-TBST. The enzyme-linked immunosorbent assays (ELISAs) were developed with the 3,3′,5,5′-tetramethylbenzidine (TMB) liquid substrate system for ELISA (T0440; Millipore Sigma). The K376-acetylated G3BP1 level was normalized with the total G3BP1 level, as determined with immunoblotting.
In vitro HDAC6 assay.
FLAG-tagged G3BP1 was immunoprecipitated from deacetylase inhibitor-treated 293T cells and eluted under native conditions with 3×FLAG peptide in TDAC assay buffer (10 mM Tris-HCl, 10 mM NaCl, pH 8.0). A sample of 1.5 μg eluted G3BP1 was treated with 0.1 μg purified recombinant His-tagged human HDAC6 (BML-SE508-0050; Enzo Life Sciences) for 1 h at 37°C and analyzed with immunoblotting.
For additional information about the reagents used, please see Table S1 in the supplemental material.
Data availability.
Subject to requirements or limitations imposed by local and/or U.S. government laws and regulations, any materials, data, and computer code originating in the authors’ laboratory relating to this study that are not available commercially and are reasonably requested by others will be made available in a timely fashion to members of the scientific community for noncommercial purposes.
Supplementary Material
ACKNOWLEDGMENTS
We thank Sidney Whiteheart (College of Medicine, University of Kentucky) and Scott Conklin (Pocono Rabbit Farm and Laboratory) for guidance on raising and purifying antibodies, Sabire Ozcan (College of Medicine, University of Kentucky) for her help with quantitative PCR and ELISA, and James Geddes (College of Medicine, University of Kentucky) and Chunaram Choudhary (Faculty of Health and Medical Sciences, University of Copenhagen) for helpful discussions.
This work was funded in part by NIH grant R01 NS077284, MDA grant MDA352743, and Department of Veteran Affairs MERIT award I01 BX002149 (to H.Z.), as well as NIH grant R21 NS095299, American Cancer Society grant IRG 85-001-25, and a Research Support Grant from the Office of the Vice President for Research of the University of Kentucky (to J.G.).
J.G. and H.Z. conceived and planned the experiments. J.G., J.C., D.-Y.N., L.T., and K.R.B carried out the experiments. J.G., J.C., L.T., and H.Z. contributed to the analysis and interpretation of the results. J.G. and H.Z. wrote and edited the manuscript. All authors provided critical feedback on the manuscript.
We have no conflicts of interest to declare relevant to this study.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00052-19.
REFERENCES
- 1.Anderson P, Kedersha N. 2008. Stress granules: the Tao of RNA triage. Trends Biochem Sci 33:141–150. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 2.Aulas A, Vande Velde C. 2015. Alterations in stress granule dynamics driven by TDP-43 and FUS: a link to pathological inclusions in ALS? Front Cell Neurosci 9:423. doi: 10.3389/fncel.2015.00423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parker F, Maurier F, Delumeau I, Duchesne M, Faucher D, Debussche L, Dugue A, Schweighoffer F, Tocque B. 1996. A Ras-GTPase-activating protein SH3-domain-binding protein. Mol Cell Biol 16:2561–2569. doi: 10.1128/mcb.16.6.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tourriere H, Chebli K, Zekri L, Courselaud B, Blanchard JM, Bertrand E, Tazi J. 2003. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J Cell Biol 160:823–831. doi: 10.1083/jcb.200212128. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Aulas A, Stabile S, Vande Velde C. 2012. Endogenous TDP-43, but not FUS, contributes to stress granule assembly via G3BP. Mol Neurodegener 7:54. doi: 10.1186/1750-1326-7-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tourriere H, Gallouzi IE, Chebli K, Capony JP, Mouaikel J, van der Geer P, Tazi J. 2001. RasGAP-associated endoribonuclease G3Bp: selective RNA degradation and phosphorylation-dependent localization. Mol Cell Biol 21:7747–7760. doi: 10.1128/MCB.21.22.7747-7760.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gallouzi IE, Parker F, Chebli K, Maurier F, Labourier E, Barlat I, Capony JP, Tocque B, Tazi J. 1998. A novel phosphorylation-dependent RNase activity of GAP-SH3 binding protein: a potential link between signal transduction and RNA stability. Mol Cell Biol 18:3956–3965. doi: 10.1128/mcb.18.7.3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bikkavilli RK, Malbon CC. 2011. Arginine methylation of G3BP1 in response to Wnt3a regulates beta-catenin mRNA. J Cell Sci 124:2310–2320. doi: 10.1242/jcs.084046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ortega AD, Willers IM, Sala S, Cuezva JM. 2010. Human G3BP1 interacts with beta-F1-ATPase mRNA and inhibits its translation. J Cell Sci 123:2685–2696. doi: 10.1242/jcs.065920. [DOI] [PubMed] [Google Scholar]
- 10.Atlas R, Behar L, Sapoznik S, Ginzburg I. 2007. Dynamic association with polysomes during P19 neuronal differentiation and an untranslated-region-dependent translation regulation of the tau mRNA by the tau mRNA-associated proteins IMP1, HuD, and G3BP1. J Neurosci Res 85:173–183. doi: 10.1002/jnr.21099. [DOI] [PubMed] [Google Scholar]
- 11.Reineke LC, Tsai WC, Jain A, Kaelber JT, Jung SY, Lloyd RE. 2017. Casein kinase 2 is linked to stress granule dynamics through phosphorylation of the stress granule nucleating protein G3BP1. Mol Cell Biol 37:e00596-16. doi: 10.1128/MCB.00596-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kedersha N, Panas MD, Achorn CA, Lyons S, Tisdale S, Hickman T, Thomas M, Lieberman J, McInerney GM, Ivanov P, Anderson P. 2016. G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J Cell Biol 212:845–860. doi: 10.1083/jcb.201508028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsai WC, Gayatri S, Reineke LC, Sbardella G, Bedford MT, Lloyd RE. 2016. Arginine demethylation of G3BP1 promotes stress granule assembly. J Biol Chem 291:22671–22685. doi: 10.1074/jbc.M116.739573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. 2009. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 15.Kwon S, Zhang Y, Matthias P. 2007. The deacetylase HDAC6 is a novel critical component of stress granules involved in the stress response. Genes Dev 21:3381–3394. doi: 10.1101/gad.461107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. 1996. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 17.Weinert BT, Narita T, Satpathy S, Srinivasan B, Hansen BK, Scholz C, Hamilton WB, Zucconi BE, Wang WW, Liu WR, Brickman JM, Kesicki EA, Lai A, Bromberg KD, Cole PA, Choudhary C. 2018. Time-resolved analysis reveals rapid dynamics and broad scope of the CBP/p300 acetylome. Cell 174:231. doi: 10.1016/j.cell.2018.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maris C, Dominguez C, Allain FH. 2005. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J 272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 19.Atlas R, Behar L, Elliott E, Ginzburg I. 2004. The insulin-like growth factor mRNA binding-protein IMP-1 and the Ras-regulatory protein G3BP associate with tau mRNA and HuD protein in differentiated P19 neuronal cells. J Neurochem 89:613–626. doi: 10.1111/j.1471-4159.2004.02371.x. [DOI] [PubMed] [Google Scholar]
- 20.Gal J, Kuang L, Barnett KR, Zhu BZ, Shissler SC, Korotkov KV, Hayward LJ, Kasarskis EJ, Zhu H. 2016. ALS mutant SOD1 interacts with G3BP1 and affects stress granule dynamics. Acta Neuropathol 132:563–576. doi: 10.1007/s00401-016-1601-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neumann H, Peak-Chew SY, Chin JW. 2008. Genetically encoding N(epsilon)-acetyllysine in recombinant proteins. Nat Chem Biol 4:232–234. doi: 10.1038/nchembio.73. [DOI] [PubMed] [Google Scholar]
- 22.Neumann H, Hancock SM, Buning R, Routh A, Chapman L, Somers J, Owen-Hughes T, van Noort J, Rhodes D, Chin JW. 2009. A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylation. Mol Cell 36:153–163. doi: 10.1016/j.molcel.2009.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuki H, Takahashi M, Higuchi M, Makokha GN, Oie M, Fujii M. 2013. Both G3BP1 and G3BP2 contribute to stress granule formation. Genes Cells 18:135–146. doi: 10.1111/gtc.12023. [DOI] [PubMed] [Google Scholar]
- 24.Soncini C, Berdo I, Draetta G. 2001. Ras-GAP SH3 domain binding protein (G3BP) is a modulator of USP10, a novel human ubiquitin specific protease. Oncogene 20:3869–3879. doi: 10.1038/sj.onc.1204553. [DOI] [PubMed] [Google Scholar]
- 25.Sowa ME, Bennett EJ, Gygi SP, Harper JW. 2009. Defining the human deubiquitinating enzyme interaction landscape. Cell 138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Solomon S, Xu Y, Wang B, David MD, Schubert P, Kennedy D, Schrader JW. 2007. Distinct structural features of caprin-1 mediate its interaction with G3BP-1 and its induction of phosphorylation of eukaryotic translation initiation factor 2alpha, entry to cytoplasmic stress granules, and selective interaction with a subset of mRNAs. Mol Cell Biol 27:2324–2342. doi: 10.1128/MCB.02300-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kedersha N, Anderson P. 2007. Mammalian stress granules and processing bodies. Methods Enzymol 431:61–81. doi: 10.1016/S0076-6879(07)31005-7. [DOI] [PubMed] [Google Scholar]
- 28.Lasko LM, Jakob CG, Edalji RP, Qiu W, Montgomery D, Digiammarino EL, Hansen TM, Risi RM, Frey R, Manaves V, Shaw B, Algire M, Hessler P, Lam LT, Uziel T, Faivre E, Ferguson D, Buchanan FG, Martin RL, Torrent M, Chiang GG, Karukurichi K, Langston JW, Weinert BT, Choudhary C, de Vries P, Van Drie JH, McElligott D, Kesicki E, Marmorstein R, Sun C, Cole PA, Rosenberg SH, Michaelides MR, Lai A, Bromberg KD. 2017. Discovery of a selective catalytic p300/CBP inhibitor that targets lineage-specific tumours. Nature 550:128–132. doi: 10.1038/nature24028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fredriksson S, Gullberg M, Jarvius J, Olsson C, Pietras K, Gustafsdottir SM, Ostman A, Landegren U. 2002. Protein detection using proximity-dependent DNA ligation assays. Nat Biotechnol 20:473–477. doi: 10.1038/nbt0502-473. [DOI] [PubMed] [Google Scholar]
- 30.Weinert BT, Scholz C, Wagner SA, Iesmantavicius V, Su D, Daniel JA, Choudhary C. 2013. Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Rep 4:842–851. doi: 10.1016/j.celrep.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 31.Scholz C, Weinert BT, Wagner SA, Beli P, Miyake Y, Qi J, Jensen LJ, Streicher W, McCarthy AR, Westwood NJ, Lain S, Cox J, Matthias P, Mann M, Bradner JE, Choudhary C. 2015. Acetylation site specificities of lysine deacetylase inhibitors in human cells. Nat Biotechnol 33:415–423. doi: 10.1038/nbt.3130. [DOI] [PubMed] [Google Scholar]
- 32.Mertins P, Qiao JW, Patel J, Udeshi ND, Clauser KR, Mani DR, Burgess MW, Gillette MA, Jaffe JD, Carr SA. 2013. Integrated proteomic analysis of post-translational modifications by serial enrichment. Nat Methods 10:634–637. doi: 10.1038/nmeth.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haggarty SJ, Koeller KM, Wong JC, Grozinger CM, Schreiber SL. 2003. Domain-selective small-molecule inhibitor of histone deacetylase 6 (HDAC6)-mediated tubulin deacetylation. Proc Natl Acad Sci U S A 100:4389–4394. doi: 10.1073/pnas.0430973100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li YR, King OD, Shorter J, Gitler AD. 2013. Stress granules as crucibles of ALS pathogenesis. J Cell Biol 201:361–372. doi: 10.1083/jcb.201302044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baron DM, Kaushansky LJ, Ward CL, Sama RR, Chian RJ, Boggio KJ, Quaresma AJ, Nickerson JA, Bosco DA. 2013. Amyotrophic lateral sclerosis-linked FUS/TLS alters stress granule assembly and dynamics. Mol Neurodegener 8:30. doi: 10.1186/1750-1326-8-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu-Yesucevitz L, Bilgutay A, Zhang Y-J, Vanderweyde T, Vanderwyde T, Citro A, Mehta T, Zaarur N, McKee A, Bowser R, Sherman M, Petrucelli L, Wolozin B. 2010. Tar DNA binding protein-43 (TDP-43) associates with stress granules: analysis of cultured cells and pathological brain tissue. PLoS One 5:e13250. doi: 10.1371/journal.pone.0013250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gal J, Zhang J, Kwinter DM, Zhai J, Jia H, Jia J, Zhu H. 2011. Nuclear localization sequence of FUS and induction of stress granules by ALS mutants. Neurobiol Aging 32:2323.e27–e2323.e40. doi: 10.1016/j.neurobiolaging.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bosco DA, Lemay N, Ko HK, Zhou H, Burke C, Kwiatkowski TJ Jr, Sapp P, McKenna-Yasek D, Brown RH Jr, Hayward LJ. 2010. Mutant FUS proteins that cause amyotrophic lateral sclerosis incorporate into stress granules. Hum Mol Genet 19:4160–4175. doi: 10.1093/hmg/ddq335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dammer EB, Fallini C, Gozal YM, Duong DM, Rossoll W, Xu P, Lah JJ, Levey AI, Peng J, Bassell GJ, Seyfried NT. 2012. Coaggregation of RNA-binding proteins in a model of TDP-43 proteinopathy with selective RGG motif methylation and a role for RRM1 ubiquitination. PLoS One 7:e38658. doi: 10.1371/journal.pone.0038658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Repici M, Hassanjani M, Maddison DC, Garcao P, Cimini S, Patel B, Szego EM, Straatman KR, Lilley KS, Borsello T, Outeiro TF, Panman L, Giorgini F. 2018. The Parkinson’s disease-linked protein DJ-1 associates with cytoplasmic mRNP granules during stress and neurodegeneration. Mol Neurobiol 56:61–77. doi: 10.1007/s12035-018-1084-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vanderweyde T, Yu H, Varnum M, Liu-Yesucevitz L, Citro A, Ikezu T, Duff K, Wolozin B. 2012. Contrasting pathology of the stress granule proteins TIA-1 and G3BP in tauopathies. J Neurosci 32:8270–8283. doi: 10.1523/JNEUROSCI.1592-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sahoo PK, Lee SJ, Jaiswal PB, Alber S, Kar AN, Miller-Randolph S, Taylor EE, Smith T, Singh B, Ho TS, Urisman A, Chand S, Pena EA, Burlingame AL, Woolf CJ, Fainzilber M, English AW, Twiss JL. 2018. Axonal G3BP1 stress granule protein limits axonal mRNA translation and nerve regeneration. Nat Commun 9:3358. doi: 10.1038/s41467-018-05647-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.d'Ydewalle C, Bogaert E, Van Den Bosch L. 2012. HDAC6 at the intersection of neuroprotection and neurodegeneration. Traffic 13:771–779. doi: 10.1111/j.1600-0854.2012.01347.x. [DOI] [PubMed] [Google Scholar]
- 44.Gal J, Chen J, Barnett KR, Yang L, Brumley E, Zhu H. 2013. HDAC6 regulates mutant SOD1 aggregation through two SMIR motifs and tubulin acetylation. J Biol Chem 288:15035–15045. doi: 10.1074/jbc.M112.431957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kawaguchi Y, Kovacs JJ, McLaurin A, Vance JM, Ito A, Yao TP. 2003. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell 115:727–738. doi: 10.1016/s0092-8674(03)00939-5. [DOI] [PubMed] [Google Scholar]
- 46.Batchu SN, Brijmohan AS, Advani A. 2016. The therapeutic hope for HDAC6 inhibitors in malignancy and chronic disease. Clin Sci (Lond) 130:987–1003. doi: 10.1042/CS20160084. [DOI] [PubMed] [Google Scholar]
- 47.Benoy V, Vanden Berghe P, Jarpe M, Van Damme P, Robberecht W, Van Den Bosch L. 2017. Development of improved HDAC6 inhibitors as pharmacological therapy for axonal Charcot-Marie-Tooth disease. Neurotherapeutics 14:417–428. doi: 10.1007/s13311-016-0501-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guedes-Dias P, de Proenca J, Soares TR, Leitao-Rocha A, Pinho BR, Duchen MR, Oliveira JM. 2015. HDAC6 inhibition induces mitochondrial fusion, autophagic flux and reduces diffuse mutant huntingtin in striatal neurons. Biochim Biophys Acta 1852:2484–2493. doi: 10.1016/j.bbadis.2015.08.012. [DOI] [PubMed] [Google Scholar]
- 49.Butler KV, Kalin J, Brochier C, Vistoli G, Langley B, Kozikowski AP. 2010. Rational design and simple chemistry yield a superior, neuroprotective HDAC6 inhibitor, tubastatin A. J Am Chem Soc 132:10842–10846. doi: 10.1021/ja102758v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li G, Jiang H, Chang M, Xie H, Hu L. 2011. HDAC6 alpha-tubulin deacetylase: a potential therapeutic target in neurodegenerative diseases. J Neurol Sci 304:1–8. doi: 10.1016/j.jns.2011.02.017. [DOI] [PubMed] [Google Scholar]
- 51.d’Ydewalle C, Krishnan J, Chiheb DM, Van Damme P, Irobi J, Kozikowski AP, Vanden Berghe P, Timmerman V, Robberecht W, Van Den Bosch L. 2011. HDAC6 inhibitors reverse axonal loss in a mouse model of mutant HSPB1-induced Charcot-Marie-Tooth disease. Nat Med 17:968–974. doi: 10.1038/nm.2396. [DOI] [PubMed] [Google Scholar]
- 52.Guo W, Naujock M, Fumagalli L, Vandoorne T, Baatsen P, Boon R, Ordovas L, Patel A, Welters M, Vanwelden T, Geens N, Tricot T, Benoy V, Steyaert J, Lefebvre-Omar C, Boesmans W, Jarpe M, Sterneckert J, Wegner F, Petri S, Bohl D, Vanden Berghe P, Robberecht W, Van Damme P, Verfaillie C, Van Den Bosch L. 2017. HDAC6 inhibition reverses axonal transport defects in motor neurons derived from FUS-ALS patients. Nat Commun 8:861. doi: 10.1038/s41467-017-00911-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alam U, Kennedy D. 2019. Rasputin a decade on and more promiscuous than ever? A review of G3BPs. BBA Mol Cell Res 1866:360–370. doi: 10.1016/j.bbamcr.2018.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim MM, Wiederschain D, Kennedy D, Hansen E, Yuan ZM. 2007. Modulation of p53 and MDM2 activity by novel interaction with Ras-GAP binding proteins (G3BP). Oncogene 26:4209–4215. doi: 10.1038/sj.onc.1210212. [DOI] [PubMed] [Google Scholar]
- 55.Gluzman Y, Sambrook JF, Frisque RJ. 1980. Expression of early genes of origin-defective mutants of simian virus 40. Proc Natl Acad Sci U S A 77:3898–3902. doi: 10.1073/pnas.77.7.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Evans PM, Zhang W, Chen X, Yang J, Bhakat KK, Liu C. 2007. Kruppel-like factor 4 is acetylated by p300 and regulates gene transcription via modulation of histone acetylation. J Biol Chem 282:33994–34002. doi: 10.1074/jbc.M701847200. [DOI] [PubMed] [Google Scholar]
- 57.Evans PM, Chen X, Zhang W, Liu C. 2010. KLF4 interacts with beta-catenin/TCF4 and blocks p300/CBP recruitment by beta-catenin. Mol Cell Biol 30:372–381. doi: 10.1128/MCB.00063-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cashman NR, Durham HD, Blusztajn JK, Oda K, Tabira T, Shaw IT, Dahrouge S, Antel JP. 1992. Neuroblastoma x spinal cord (NSC) hybrid cell lines resemble developing motor neurons. Dev Dyn 194:209–221. doi: 10.1002/aja.1001940306. [DOI] [PubMed] [Google Scholar]
- 59.Durham HD, Dahrouge S, Cashman NR. 1993. Evaluation of the spinal cord neuron X neuroblastoma hybrid cell line NSC-34 as a model for neurotoxicity testing. Neurotoxicology 14:387–395. [PubMed] [Google Scholar]
- 60.Heinzel SS, Krysan PJ, Calos MP, DuBridge RB. 1988. Use of simian virus 40 replication to amplify Epstein-Barr virus shuttle vectors in human cells. J Virol 62:3738–3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gao YS, Hubbert CC, Lu J, Lee YS, Lee JY, Yao TP. 2007. Histone deacetylase 6 regulates growth factor-induced actin remodeling and endocytosis. Mol Cell Biol 27:8637–8647. doi: 10.1128/MCB.00393-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kamelgarn M, Chen J, Kuang L, Arenas A, Zhai J, Zhu H, Gal J. 2016. Proteomic analysis of FUS interacting proteins provides insights into FUS function and its role in ALS. Biochim Biophys Acta 1862:2004–2014. doi: 10.1016/j.bbadis.2016.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.UniProt Consortium. 2015. UniProt: a hub for protein information. Nucleic Acids Res 43:D204–212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lv Y, Zhao SG, Lu G, Leung CK, Xiong ZQ, Su XW, Ma JL, Chan WY, Liu HB. 2017. Identification of reference genes for qRT-PCR in granulosa cells of healthy women and polycystic ovarian syndrome patients. Sci Rep 7:6961. doi: 10.1038/s41598-017-07346-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gibson CJ, Hossain MM, Richardson JR, Aleksunes LM. 2012. Inflammatory regulation of ATP binding cassette efflux transporter expression and function in microglia. J Pharmacol Exp Ther 343:650–660. doi: 10.1124/jpet.112.196543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Buchan JR, Nissan T, Parker R. 2010. Analyzing P-bodies and stress granules in Saccharomyces cerevisiae. Methods Enzymol 470:619–640. doi: 10.1016/S0076-6879(10)70025-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Subject to requirements or limitations imposed by local and/or U.S. government laws and regulations, any materials, data, and computer code originating in the authors’ laboratory relating to this study that are not available commercially and are reasonably requested by others will be made available in a timely fashion to members of the scientific community for noncommercial purposes.