Fig. 7.
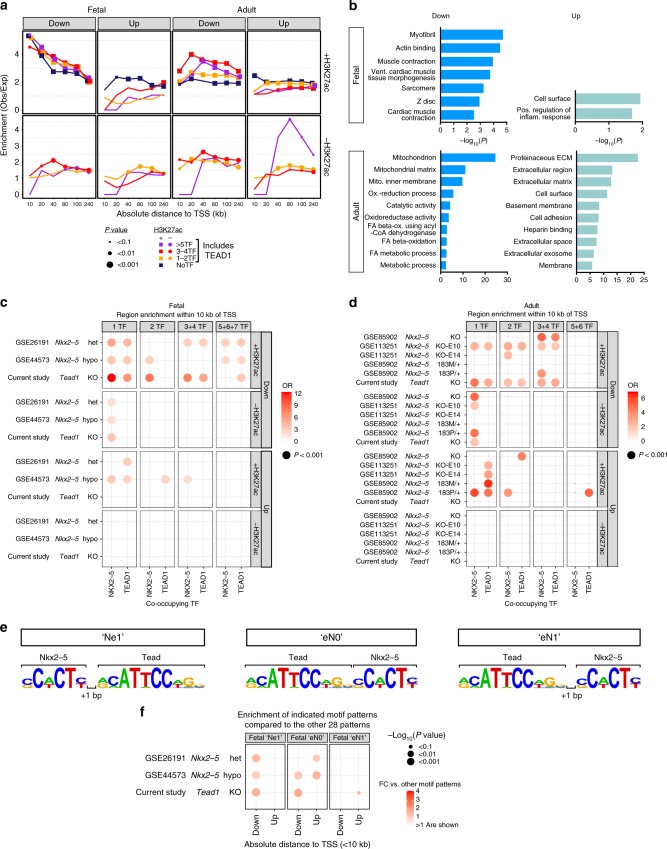
TEAD1 is an integral regulator of cardiac gene expression. a Enrichment of TF or H3K27ac regions adjacent to genes up-regulated and down-regulated in fetal or adult Tead1 knockout hearts. TF regions contained TEAD1. Enrichment (observed/expected) of TF or H3K27ac regions within the indicated distance of differentially expressed genes’ TSS was calculated by permutation testing. Expected values were determined by randomly distributing regions across the mappable genome in 3000 permutations. Size of symbols indicate permutation test P-value. TEAD1-occupied regions marked by H3K27ac were enriched adjacent to upregulated DEGs. b GO term analysis of genes down-regulated or up-regulated in TEAD1 knockout and adjacent to TEAD1-bound regions. Down-regulated, TEAD1-bound genes were enriched for biological process terms related to cardiomyocyte-specific functions. c, d Bubble plots indicating enrichment of regions occupied by the indicated TF (NKX2-5 or TEAD1) within 10 kb of the TSS of up-regulated or down-regulated genes in the indicated datasets involving either Tead1 knockout or Nkx2-5 mutation. NKX2-5 or TEAD1-bound regions marked by H3K27ac were enriched adjacent to genes downregulated by Nkx2-5 or Tead1 mutation. e “Preferred” arrangements of NKX2-5 and TEAD1 motifs in regions co-bound by these TFs (see Supplementary Fig. 10j). f Enrichment of regions co-bound by NKX2-5 and TEAD1 and containing the indicated motif arrangement adjacent to differentially expressed genes in fetal heart. Bubble size indicates permutation P-value (3000 permutations). Color indicates fold-change of enrichment compared to co-bound regions containing the 28 non-preferred motif arrangements. Source data for panels a, c, d, f are provided as a Source Data file