Figure 2.
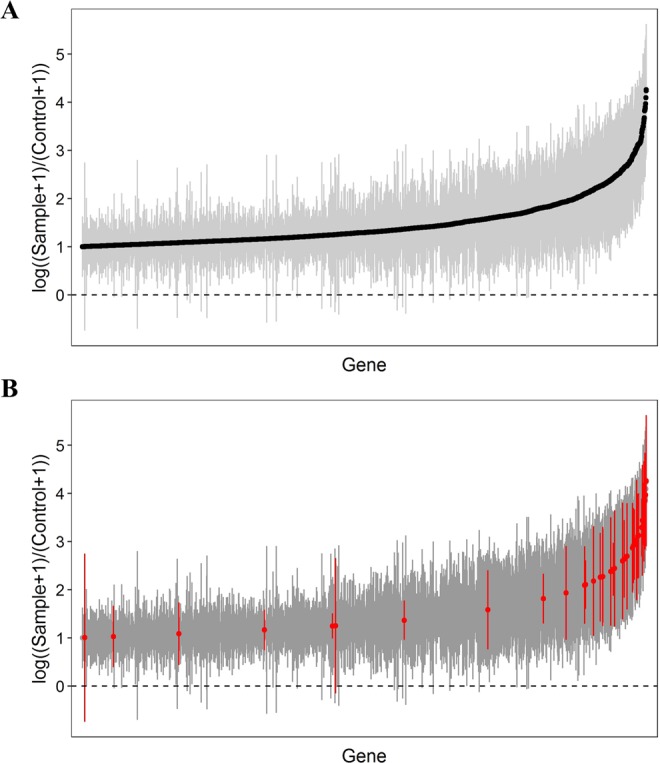
shRNA screening data after applying a strict cut-off. (A) shRNA screening data were filtered by establishing three criteria: mean ratio higher than 1 (Log Ratio >1) compared to the uninfected control, standard error lower than 2 (SE <2) and a consistency higher than 60%. (B) Out of 2,888 genes, twenty-nine host genes were selected for further validation. High effect in the shRNA screening after MRSA infection and/or a biological relevance for intracellular pathogenesis were used as criteria to choose the most interesting hits. Selected genes are highlighted in red.
