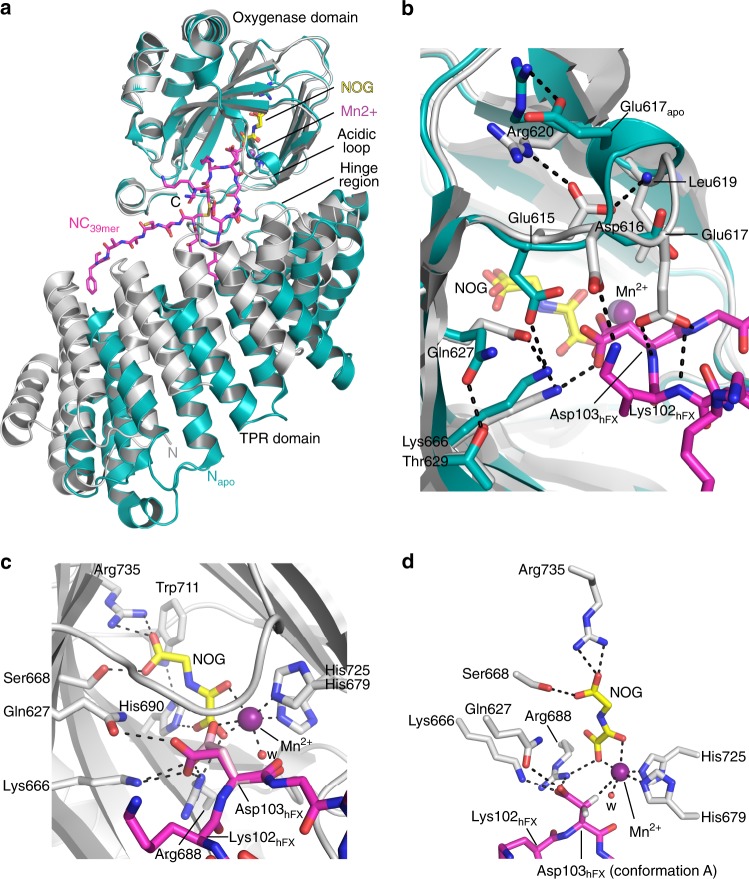
Fig. 4.
Significant conformational changes occur in AspH upon substrate binding. Color code: grey: His6-AspH315–758 (AspH-TPR-Ox:hFX); turquoise: His6-AspH315–758 (AspH-TPR-Ox); magenta: carbon-backbone of NC39mer peptide; yellow: carbon-backbone of N-oxalylglycine (NOG); violet: Mn; red: oxygen; blue: nitrogen; pale yellow: sulfur. w: water. a Superimposition of the AspH-TPR-Ox:hFX (grey) and AspH-TPR-Ox (turquoise) structures indicate conformational changes in the TPR domain, the hinge region, and the oxygenase acidic loop on substrate binding. b Superimposition of the AspH-active sites of the AspH-TPR-Ox:hFX (grey) and AspH-TPR-Ox (turquoise) structures: The interaction between Glu617apo and Arg620 (2.5 Å) in the AspH-TPR-Ox structure is lost on substrate binding; both Asp616 and Glu617 interact with the substrate in the AspH-TPR-Ox:hFX structure. Gln627 (3.2 Å) and Lys666 (2.7 Å) bind to the Asp103hFX carboxylate of the active conformer of the AspH-substrate. On substrate binding, the side chain of Glu615 rotates by ~90° to interact with the side chain of Arg620 (2.8 Å) and the main chain of Leu619 (2.8 Å), rather than Lys666 (2.7 Å) as in the substrate unbound state. c The side chain of the Asp103hFX residue undergoing hydroxylation is observed in two conformations (A: magenta and B: pink; see Supplementary Fig. 10 for details). The Asp103hFX side chain carboxylate of conformation B (pink) is positioned (2.6 Å) to interact with the Mn. d Close-up of the AspH-active site: The pro-R hydrogen at the Asp103hFX β-position of the likely productive NC39mer conformation A (magenta) is positioned to interact with the Mn (distance Cβ-Mn: 4.2 Å), consistent with hydroxylation at this position