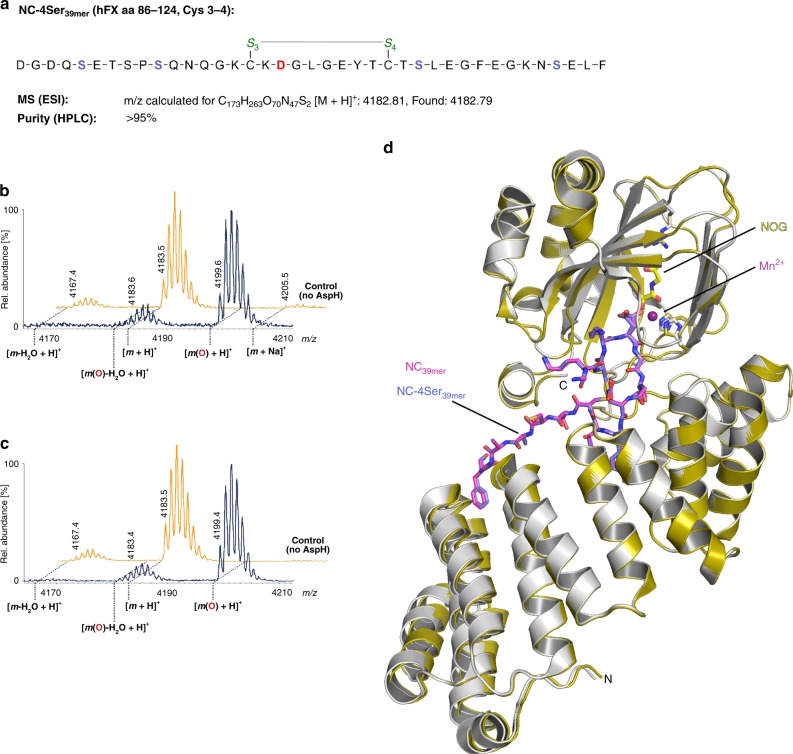
Fig. 8.
The AspH-substrate requirement is a ring composed of 10 amino acid residues. End-point turnover reactions were performed as in the Methods Section. Light orange graphs represent controls in which AspH was replaced by buffer. a Schematic structure and calculated mass of the NC-4Ser39mer peptide featuring a single disulfide between Cys3–4 (green); its sequence is identical to the one of hFX EGF139mer (see Fig. 1a), except that Cys90hFX, 95, 112, 121 are substituted for Ser (light blue) to avoid disulfide scrambling; the hydroxylation site (Asp103hFX) is in red. b >95% Hydroxylation was observed under standard (non-redox) conditions. c >95% Hydroxylation was observed under redox conditions. d Superimposition of the AspH-TPR-Ox:hFX crystal structure (color code: AspH: grey; NC39mer peptide: magenta) with the AspH-TPR-Ox:NC-Ser39mer crystal structure (color code: AspH: gold; NC-4Ser39mer peptide: slate blue) shows a high conservation of the conformations of both enzyme and ligands