Abstract
Objective:
To identify differences in gene expression between patients with in-hospital delirium from a known etiology (urinary tract infection [UTI]) and patients with delirium from an unknown etiology, as well as from nondelirious patients.
Methods:
Thirty patients with delirium (8 with UTI) and 21 nondelirious patients (11 with UTI) were included in this prospective case–control study. Transcriptomic profiles from messenger RNA sequencing of peripheral blood were analyzed for gene expression and disease-specific pathway enrichment patterns, correcting for systemic inflammatory response syndrome. Genes and pathways with significant differential activity based on Fisher exact test (P < .05, |Zscore| >2) are reported.
Results:
Patients with delirium with UTI, compared to patients with delirium without UTI, exhibited significant activation of interferon signaling, upstream cytokines, and transcription regulators, as well as significant inhibition of actin cytoskeleton, integrin, paxillin, glioma invasiveness signaling, and upstream growth factors. All patients with delirium, compared to nondelirious patients, had significant complement system activation. Among patients with delirium without UTI, compared to nondelirious patients without UTI, there was significant activation of elF4 and p7056 K signaling.
Conclusions:
Differences exist in gene expression between delirious patients due to UTI presence, as well as due to the presence of delirium alone. Transcriptional profiling may help develop etiology-specific biomarkers for patients with delirium.
Keywords: delirium, urinary tract infection, transcriptome, RNA sequencing
Introduction
Delirium is an acute cognitive disorder characterized by fluctuations in attention and awareness. Approximately 18% to 35% of elderly patients present to the hospital with delirium and 10% to 15% develop delirium during hospitalization, with increased incidences among critically ill patients.1–3 Delirium is associated with increased health-care utilization leading to greater hospitalization costs,4 longer hospital stays,4,5 and worse clinical outcomes.5–7
Identification of delirium’s cause is imperative for its treatment. There are numerous factors that increase delirium risk, including advanced age5,8; baseline cognitive,9 hearing, or vision impairments2,10,11; presence of infection12; and polypharmacy.13,14 Patients suspected of having delirium often undergo extensive testing of serum, urine, and cerebrospinal fluid; electroencephalography; and neuroimaging.3 However, such an indiscriminate workup is inefficient and costly, and despite this, the cause of delirium in many patients remains unknown.
One method for classifying diseases by type and pathogenesis is through identification of unique gene expression signatures.15–17 RNA sequencing (RNA-Seq)18 uses next-generation sequencing technologies to efficiently quantify gene transcripts in a biological sample across the whole human genome with high fidelity.19 While previous studies focused on a narrow set of inflammatory markers that are believed a priori to play a role in delirium pathophysiology, profiling the whole transcriptome allows for a more unbiased approach in identifying particular genes and pathways that are differentially up-or downregulated in delirium.20–22 Our study is the first to implement RNA-Seq in delirium research. We hypothesized that patients with in-hospital delirium associated with a urinary tract infection (UTI) will have differences in mRNA gene expression in comparison with patients with delirium due to other causes, as well as compared to patients hospitalized without delirium, with or without an UTI.
Methods
Study Participants
This prospective case–control study took place from July 2014 through March 2015. Patients were identified on the neurology or medicine services at a tertiary referral hospital; patients with delirium or UTI were eligible for inclusion. All patients admitted to the neurology and medicine services were prospectively screened in the electronic medical record daily (H.A.S., V.C.D.) for altered mental status and/or presence of UTI. The primary physician caring for the patient was contacted to determine whether or not the patient met the inclusion criteria. If the patient met the inclusion criteria, H.A.S. approached him or her for consent, specimen collection, and a brief interview. Delirium was confirmed using the confusion assessment method by a board-certified neurologist (V.C.D.).23 Blood samples were collected from cases within 24 hours of delirium diagnosis in all but 2, where blood was obtained within 48 hours of diagnosis. Delirium was attributed to the UTI when the 2 conditions occurred contemporaneously and when there was no other more likely cause of the delirium. Urinary tract infection was defined as a urinalysis with the presence of white cells and a urine culture with greater than 100 000 colony-forming units of an infectious organism. Patients cared for by a surgical service were excluded. Information about cognitive impairment, mobility, hearing, and vision was obtained from the patient or an informant when possible. Informants were asked about baseline cognitive function using the IQCode.24 In cases where the informant was not available and the patient could not provide the information, these data were obtained from chart review. Statistical comparisons between the 2 groups were calculated using a 2-sample unequal variance t test.
Standard Protocol Approvals, Registrations, and Patient Consents
We obtained consent for study enrollment from each patient or the patient’s surrogate decision maker. Eligible patients were enrolled under a research protocol (#13–12557) approved by the institutional review board of the University of California, San Francisco.
Peripheral Blood Mononuclear Cell Isolation
Patient blood samples were collected in tubes with a preservative solution containing trisodium citrate, citric acid, and dextrose.
Tubes were centrifuged, and the plasma fraction was removed. The blood sample was diluted to a 1:1 ratio with phosphate-buffered saline (PBS). Diluted blood was layered onto 10 mL of Ficoll mixture and centrifuged. After removing the top layer, the cells in the buffy coat were transferred, washed with PBS, counted, spun down, and stored in liquid nitrogen.
RNA Sequencing Library Preparation
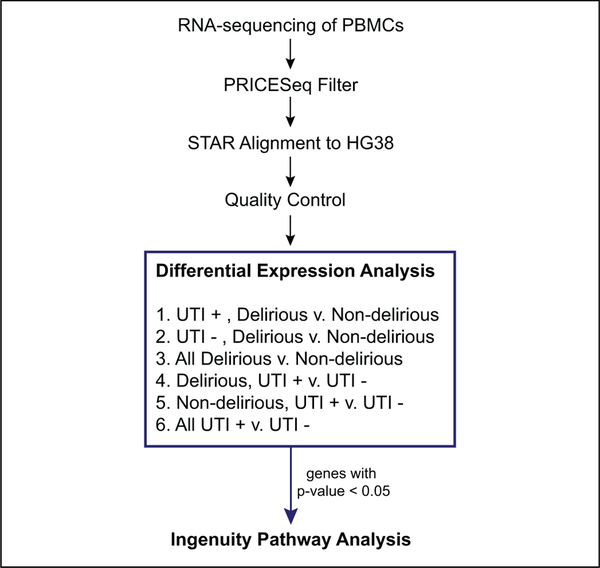
RNA was extracted from each peripheral blood mononuclear cell sample and was depleted of ribosomal RNA using Ribo-Zero, (Illumina, San Diego, California CA) (Figure 1). The Illumina TruSeq, (Illumina, San Diego, CA) protocol was utilized to generate RNA-Seq libraries from 1 to 2 ng of RNA/sample. Samples were individually barcoded, allowing for the libraries to be pooled and sequenced together on an Illumina HiSeq 4000 machine.
Figure 1.
First, RNA was extracted from PBMCs and sequenced on Illumina HiSeq 2500. The .fastq files obtained from sequencing were quality filtered using PRICESeqFilter and subsequently aligned to HG38 using STAR aligner. After alignment, quality control filters included removal of samples with less than 60% uniquely mapped reads, removal of nonprotein-coding genes, removal of ribosomal and mitochondrial rRNA, and removal of genes with zero read counts in greater than 75% of samples. After quality control, differential expression analysis was performed using DESeq2 on each of the 6 cross sections of the data shown. Genes with raw P value < .05, for each comparison, were included in ingenuity pathways analysis. PBMC indicates peripheral blood mononuclear cell; rRNA, ribosomal ribo-nucleic acid STAR, spliced transcripts alignment to reference.
Transcript Alignment and Quality Filtering
Reads were quality filtered using PriceSeqFilter25 and then aligned to the human genome (NCBI GRCh-38) using the spliced transcripts alignment to reference (STAR) aligner.26 Summing all samples together, there were 542 251 466 total single-end 50 base-pair reads. Samples with less than 50% uniquely mapping reads were discarded from the analysis. Abundance of aligned reads mapping to transcripts was estimated by STAR. Transcript counts were filtered to include only genes listed with biotype “protein-coding” in the ENSEMBL database.27 Mitochondrial and ribosomal RNA genes were removed along with genes with zero-mapped reads in ≥75% of samples to reduce spurious differential expression results.
Differential Expression Analysis
Differential expression analysis was performed using DESeq228 on cross sections of the patient data to understand how each of the conditions impacted the disease phenotype. In a preliminary analysis, differential expression was performed without correcting for potential confounding variables. However, after observing upregulation of numerous inflammatory markers, we added a correction for systemic inflammatory response syndrome (SIRS) by including a binary variable for SIRS status in the DESeq2 model for differential expression. Systemic inflammatory response syndrome status was determined at the time of study enrollment based on standard SIRS criteria,29 determined by meeting 2 or more of the following: temperature greater than 38°C or less than 36°C, heart rate greater than 90 beats per minute, respiratory rate greater than 20 breaths per minute, and white blood count greater than 12 000 or less than 4000. We calculated P values and adjusted P values using Benjamini-Hochberg multiple test correction to determine the false discovery rate.30 However, given that fewer than 10 genes were significant at Padj < .05 across all comparisons, all genes with unadjusted P value <.05 were selected as inputs to the pathway functional enrichment analysis as described below.
Pathway Functional Enrichment Analysis
GO enrichment analysis was performed using the ToppGene enrichment analysis tool31 to identify biological processes overrepresented in the list of differentially expressed genes with an unadjusted P < .05. Qiagen’s Ingenuity Pathway Analysis (IPA) software (QIAGEN, Redwood City, California) was used to further understand the activation and repression of specific pathways. Genes with unadjusted P < .05 were input into the core analysis function using the Ingenuity database as the null model. Ingenuity Pathway Analysis–canonical path-way analysis aims to identify pathways that are significantly over-or underrepresented, while the IPA upstream regulator analysis seeks to identify the upstream transcriptional regulators that can explain the observed gene expression.32 Results from the canonical pathways and upstream regulator analysis were interpreted with respect to the existing literature on delirium. The software computed a Fisher exact test P value, which was not corrected for multiple testing. Ingenuity Pathway Analysis provides 2 metrics for evaluation of the significance of results: the overlap P value and z score. The overlap P value, calculated as a Fisher exact test, indicates the enrichment of genes in a pathway without taking into account the regulation direction. Meanwhile, the z score assesses the correlation of observed and predicted up-/downregulation patterns. Analysis of these results focused only on pathways that were both significant at unadjusted P < .05 and |z score| > 2.
Hierarchical Clustering of Transcriptome Profile
Genes differentially expressed in the comparison of all delirious versus nondelirious patients at unadjusted P < .01 were used as features for hierarchical clustering to demonstrate the separability between the 2 clinical classifications based on their transcriptome profiles. The rlog transformation from the DESeq2 package was applied to the normalized gene counts. The data were centered and scaled prior to clustering. Clustering was performed using the hclust function in the R v 3.3.1 statistical package33 with Eucli-dean distance and complete linkage and visualized via the heat-map.2 function from the R v 3.3.1 gplots package.34
Results
Study Participants
There were 75 patients who met criteria and were approached for inclusion. Fifty one were included in the analysis, 30 with delirium and 21 without at the time of blood specimen collection. The remaining 24 patients were not included for the following reasons: refusal to participate (14), lack of a surrogate for consent (2), discharge from hospital prior to blood specimen collection (4), or low numbers of uniquely aligned reads or low numbers of unique transcripts, which are indicators of low-quality libraries during sequencing that may erroneously skew the analysis (4). Thirty hospitalized patients with delirium were included, including 8 patients with delirium attributed to UTI. Alternate causes of delirium included infection (non-UTI; 3 patients), medication toxicity (3), metabolic state (4), neoplastic state (3), postictal state (1), and a combination of categories (6). Twenty-one hospitalized controls without delirium were enrolled, 11 of whom were diagnosed with an UTI.
The mean age of patients with delirium was 69 (13) years, compared with 59 (17) years in patients without delirium (P = .04). Patients both with and without delirium were hospitalized for a broad range of primary diagnoses (Supplemental Table). Of the patients with delirium, 7 (23%) met SIRS criteria, compared with 1 (5%) patient without delirium (P = .05). Overall, patients with delirium were older and more likely to be diagnosed with dementia compared to the patients without delirium (Table 1).
Table 1.
Demographics and Clinical Characteristics of Study Participants.
| Characteristics | Delirium+, Mean (SD) |
Delirium−, Mean (SD) |
Delirium+ Versus Delirium− P Value |
||
|---|---|---|---|---|---|
| UTI+ (n = 8) | UTI− (n = 22) | UTI+ (n = 11) | UTI− (n = 10) | ||
| Age, years (SD) | 76 (5) | 66 (14) | 61 (17) | 57 (18) | .04 |
| Women, n (%) | 5 (63%) | 10 (45%) | 6 (55%) | 2 (20%) | .4 |
| Positive SIRS criteria,a n (%) | 3 (38%) | 4 (18%) | 1 (9%) | 0 (0%) | .05 |
| Dementia, n (%) | 1 (13%) | 6 (27%) | 1 (9%) | 0 (0%) | .006 |
| Mild cognitive impairment, n (%) | 3 (38%) | 3 (14%) | 2 (18%) | 0 (0%) | .3 |
| Alcohol use disorder, n (%) | 0 (0%) | 3 (14%) | 1 (9%) | 2 (20%) | .7 |
| Poor vision, n (%) | 0 (0%) | 1 (5%) | 0 (0%) | 1 (10%) | .8 |
| Hearing loss, n (%) | 2 (25%) | 12 (55%) | 3 (27%) | 2 (20%) | .2 |
| Mobility dependence, n (%) | 3 (38%) | 5 (23%) | 1 (9%) | 0 (0%) | .03 |
Abbreviations: SIRS, systemic inflammatory response syndrome; SD, standard deviation; UTI, urinary tract infection.
Positive SIRS criteria included meeting 2 or more of the following: temperature greater than 38°C or less than 36°C, heart rate greater than 90 beats per minute, respiratory rate greater than 20 breaths per minute, white blood count greater than 12 000 or less than 4000.
Analysis of Delirium of Different Etiologies
The mean percentage of uniquely aligned reads across all the participants was 79.8%, and mean total gene counts were 3.0 million. Filtering for protein coding genes reduced the mean number of gene counts per sample to 2.2 million. Differential gene expression analysis of patients with delirium resulting from an UTI versus delirium resulting from unknown etiologies identified 440 differentially expressed genes at an unadjusted P < .05. Among these differentially expressed genes, ToppGene enrichment analysis identified significant overrepresentation of genes in pathways related to response to interferon α/β signaling, platelet activation, and response to virus (Table 2).
Table 2.
Comparison of Top Canonical Pathways Between Groups After Adjusting for Systemic Inflammatory Response Syndrome.
| Group Comparison | Canonical Pathway | P Value |
z Scorea |
|---|---|---|---|
| D+ versus D− | Complement system | .007 | 2.0 |
| D+ UTI+ versus | None | n/a | n/a |
| D− UTI + | |||
| D+ UTI− versus | elF4 and p70S6K | .01 | 2 |
| D− UTI− | |||
| UTI+ versus UTI− | Interferon | .002 | 2.5 |
| Actin cytoskeleton | .00002 | −3.3 | |
| Integrin | 8.7 × I0−8 | −2.9 | |
| Paxillin | .0003 | −2.3 | |
| RhoA | .0004 | −2.3 | |
| D+ UTI+ versus | Interferon | .00004 | 2.5 |
| D+ UTI− | Actin cytoskeleton | .004 | −2.5 |
| Integrin | .002 | −2.5 | |
| Paxillin | .003 | −2.5 | |
| Glioma invasiveness | .004 | −2.2 | |
| D− UTI+ versus | None | n/a | n/a |
| D− UTI− | |||
Abbreviations: D+, delirious; D−, not delirious; n/a, not applicable; UTI+, urinary tract infection present; UTI−, urinary tract infection absent.
z score: A positive z score indicates upregulation of the pathway. A negative z score indicates downregulation of the pathway.
Interferon Signaling Is the Primary Pathway Distinguishing Delirium With UTI Versus Delirium Due To an Unknown Etiology
Compared to patients with delirium without an UTI, patients with delirium with an UTI had significant activation of the interferon signaling pathway (P = .00004, z score = 2.5) as well as notable upstream activation of cytokines and transcriptional regulators (25.8% and 22.6% of all activated upstream regulators with unadjusted P < .05 and z score > 2, respectively). Patients with delirium with an UTI had significant repression of the following signaling: integrin (P = .002, z score = −2.5), paxillin (P = .003, z score = −2.5), actin cytoskeleton (P = .004, z score = −2.5), and glioma invasiveness (P = .004, z score = −2.4). When analyzing all patients with an UTI, compared to all patients without an UTI, interferon signaling was also significant (P = .002, z score = 2.5).
Transcriptomic Profiling Separates Delirious From Nondelirious Patients
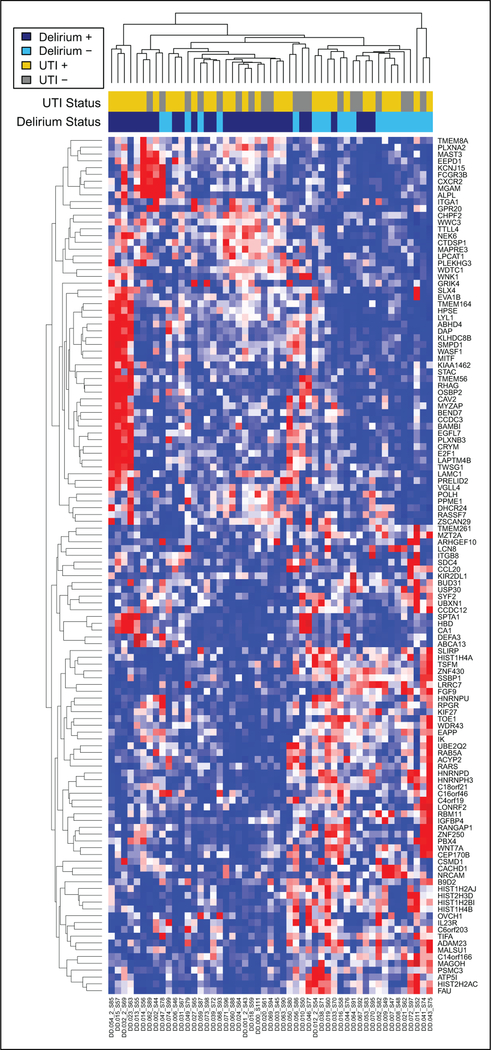
Differential expression analysis of patients with delirium of any etiology versus no delirium resulted in 670 differentially expressed genes at unadjusted P < .05, for which ToppGene enrichment analysis found overrepresentation of functional elements related to mitochondrial structure and metabolism of heme. The subset of 123 genes with unadjusted P < .01 as features, hierarchical clustering separated patients into their respective groups as seen in Figure 2.
Figure 2.
Transcriptional differences between delirious and nondelirious patients. Heatmap constructed using unsupervised hierarchical clustering (heatmap.2 R function with complete linkage and Euclidian distance) of rlog transformed, normalized expression values for genes with P value < .05 in the comparison of patients with delirium (purple, UTI+ in dark purple and UTI− in light purple) versus those without delirium (red, UTI + in dark red and UTI− in light red). UTI indicates urinary tract infection.
Pathways Associated With Differences Between Delirious and Nondelirious Patients
The 670 genes differentially expressed with P < .05 were input into IPA, and canonical pathway analysis indicated activation of complement system pathways (P = .007, z score = 2) across all samples, regardless of UTI status. When considering only the patients with UTI, no pathways were considered significantly activated or repressed. Meanwhile, when considering only the patients without UTI, regulation of eIF4 and p70S6 K signaling was upregulated in delirium (P = .01, z score = 2).
Effect of SIRS on the Activation of Interleukin-6 and Interleukin-8 Pathways
Prior to adjusting for SIRS criteria in the model for differential expression analysis between all delirious and nondelirious patients, both the interleukin (IL)-6 pathway and the IL-8 path-way were identified as activated by z score (2.1 and 2.9, respectively). However, the overlap P values were not statistically significant (0.294 and 0.236, respectively). After correction for SIRS, IL-6 was no longer identified as activated, but IL-8 was activated with a z score of 3.
Discussion
We used whole transcriptome profiling to look broadly at gene expression changes of participants with delirium due to UTI compared to those without UTI and between patients with and without delirium. The differential expression between any of these groups was subtle, with no genes being differentially expressed with an imposed adjusted P value of <.05. Thus, we relaxed this condition and used ranked unadjusted P values combined with filtering by pathway specific z scores to detect obscured or weak signals. Using this more sensitive approach, we showed that patients with delirium with an UTI, when compared to patients with delirium without an UTI, displayed significant activation of the interferon signaling pathway, upstream cytokines, and transcriptional regulators, as would be expected for infections, as well as repression of integrin, actin cytoskeleton, paxillin, and glioma invasiveness signaling. For all patients with delirium, compared to patients without delirium, there was significant activation of the complement system. When examining only patients without UTI, there was significant activation of the pathways for elF4 and p7056 K regulation in patients with delirium. These results highlight several pathways that may be implicated in distinguishing patients with delirium from those without delirium, and further, in distinguishing delirium due to specific etiologies.
Interferons are cytokines with immunomodulatory properties35 and have been associated with delirium.36,37 Our data demonstrate that interferon signaling is significantly upregulated for patients with delirium with an UTI, when compared to patients with delirium without an UTI. However, interferon signaling was not significantly upregulated, among nondelirious patients with an UTI compared to those without. This suggests that there may be a synergistic biological response when delirium is paired with infection, compared with the 2 processes in isolation. This distinction is also made in one recent prospective case–control study that included 47 patients with concomitant delirium and sepsis, 36 patients with delirium without sepsis, 36 patients with sepsis only, and 36 patients with neither delirium nor sepsis (control).36 Interferon γ was elevated in all groups except for the controls but was most elevated for patients with concurrent delirium and sepsis.
The complement pathway has been infrequently studied in patients with delirium.38 Activation of the complement system is typical of an organism’s stress response,39 and one recent study has detected activated complement in a prospective cohort study using proteomic analysis of cerebrospinal fluid in patients with delirium.39 In our investigation, there was significant activation of the complement system in patients with delirium compared to those without delirium, but this was not found in the other group comparisons. Thus, this finding is of unknown clinical significance; however, given that complement activation is a potential therapeutic target in a wide range of pathologies, further study using proteomic methods to probe for the activated forms of complement is warranted.
Activation of the cytokine IL-6 and IL-8 pathways has been associated with delirium in prior studies,22,40–43 but we were unable to replicate this finding, especially when correcting for SIRS status. Increased serum levels of IL-6 and IL-8 have been found in association with postoperative delirium in several prospective cohorts of older patients admitted for hip fracture surgery.40–42 In 1 patient, the highest levels of IL-6 were ascertained during active delirium—particularly if the delirium was classified as hyperactive or mixed, rather than hypoactive,—whereas IL-8 was highest prior to development of delirium.40 In a separate case–control study that enrolled older patients admitted for either elective or emergent surgery, a multivariate analysis of preoperative serum samples revealed that increased levels of IL-6 were associated with a greater risk of postoperative delirium.22 However, our data suggest that IL-6 and IL-8 may be more tightly correlated with a heightened inflammatory response than to delirium itself.
While RNA-Seq is new to the field of delirium research, this technique has already benefited several other areas of medicine. Patterns of peripheral blood gene expression differ in patients with pulmonary infections, distinguishing participants with a respiratory viral infection from uninfected participants, as well as delineating between viral and bacterial infection with high (93%) specificity.44 A targeted reverse transcription polymerase chain reaction assay measuring the expression of these genes was validated in a prospective emergency department cohort with 89% sensitivity and 94% specificity.45 Such an assay has the potential to increase diagnostic accuracy and bolster directed treatment. This technique is most advanced in breast oncology, where a 21-gene signature (OncoDx) classifies tumors by prognosis based on gene profiling, allowing targeted use of adjuvant chemotherapies.46,47 Our exploratory research suggests that RNA-Seq may have the ability to distinguish between different causes of delirium, and thus, further investigation using larger cohorts with equally rigorous entry criteria and phenotyping is warranted.
This investigation features several strengths. All participants were evaluated and classified by a board-certified neurologist. We corrected for SIRS in our analysis to control for a heightened inflammatory state, as many of the pathways under investigation corresponded with inflammation. Whereas prior studies were limited by the need to specify which few serum or cerebrospinal fluid markers to study, the RNA-Seq approach we employed enabled us to uncover path-ways not previously associated with delirium, albeit in an exploratory manner.
Limitations
This study is preliminary and has a few limitations. The sample size for this investigation is small. Null results may be due to both small numbers of patients and heterogeneity. An additional limitation is the issue of subject matching. Although sex was similar between delirious and nondelirious patients, patients with delirium were older and had higher rates of dementia in addition to other risk factors for delirium. Lack of a robust matching mechanism and a small sample size that precluded the possibility of multivariable modeling raise the potential for confounding by these factors. Overall, differential expression among the groups was subtle, with fewer than 10 genes differentially expressed at an adjusted P value of <.05 (Padj), and thus, we explored weaker signals by utilizing unadjusted P values and z score statistics for pathway enrichment. While we detected expected pathways using this approach, such as interferon signaling in UTI(+) versus UTI(−) patients, our approach is susceptible to false positives and is also dependent on the particular gene pathways that are defined in the current IPA database.
Since the aim of our study focused on measuring serum biomarkers at a single point in time, we did not perform longitudinal delirium assessments. Because of caregiver and provider interviews during patient identification and enrollment, it is unlikely that patients identified as nondelirious were delirious prior to study. However, it is possible that nondelirious patients developed delirium at a later time during the hospitalization following enrollment. Retrospective review of patient charts identified only 1 nondelirious patient who developed a confusional state 5 days following his blood draw; while it is unlikely that this affected our analysis, one would expect misclassification of this type to bias toward the null. Longitudinal assessment will be important for future studies, especially in measuring how gene signaling may change in association with delirium persistence and resolution.
Conclusions
There are differences in gene expression between patients with delirium due to UTI compared to those without UTI, as well as between patients with and without delirium. Whole transcriptome profiling may help differentiate between causes of delirium and shed light onto its pathophysiology. Follow-up studies using a larger cohort are needed to formally validate the use of a transcriptomic classifier of delirium etiology, but such an understanding may advance the treatment of delirium by preventing unnecessary testing.
Supplementary Material
Acknowledgments
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Supported by UCSF-RAP (M.R.W., V.C.D., H.A.S.) and National Center for Advancing Translational Sciences of the NIH (KL2TR000143; M.R.W.).
Footnotes
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Supplemental Material
Supplemental material for this article is available online.
References
- 1.Vasilevskis EE, Han JH, Hughes CG, Ely EW. Epidemiology and risk factors for delirium across hospital settings. Best Pract Res Clin Anaesthesiol 2012;26(3):277–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.LaHue SC, Liu VX. Loud and clear: sensory impairment, delirium, and functional recovery in critical illness. Am J Respir Crit Care Med 2016;194(3):252–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Inouye SK, Westendorp RG, Saczynski JS. Delirium in elderly people. Lancet. 2014;383(9920):911–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown CH, Laflam A, Max L, et al. The impact of delirium after cardiac surgical procedures on postoperative resource use. Ann Thorac Surg 2016;101(5):1663–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Francis J, Martin D, Kapoor WN. A prospective study of delirium in hospitalized elderly. JAMA. 1990;263(8):1097–1101. [PubMed] [Google Scholar]
- 6.Honda S, Nagai T, Sugano Y, et al. Prevalence, determinants, and prognostic significance of delirium in patients with acute heart failure. IntJ Cardiol 2016;222:521–527. [DOI] [PubMed] [Google Scholar]
- 7.Sato K, Kubota K, Oda H, Taniguchi T. The impact of delirium on outcomes in acute, nonintubated cardiac patients. Eur Heart J Acute Cardiovasc Care. 2017;6(6):553–559. [DOI] [PubMed] [Google Scholar]
- 8.Ryan DJ, O’Regan NA, Caoimh RO, et al. Delirium in an adult acute hospital population: predictors, prevalence, and detection. BMJ Open. 2013;3(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McNicoll L, Pisani MA, Zhang Y, Ely EW, Siegel MD, Inouye SK. Delirium in the intensive care unit: occurrence and clinical course in older patients. J Am Geriatr Soc 2003;51(5):591–598. [DOI] [PubMed] [Google Scholar]
- 10.Inouye SK. Predisposing and precipitating factors for delirium in hospitalized older patients. Dement Geriatr Cogn Disord. 1999; 10(5):393–400. [DOI] [PubMed] [Google Scholar]
- 11.Inouye SK, Zhang Y, Jones RN, Kiely DK, Yang F, Marcantonio ER. Risk factors for delirium at discharge: development and validation of a predictive model. Arch Intern Med 2007;167(13): 1406–1413. [DOI] [PubMed] [Google Scholar]
- 12.Manepalli J, Grossberg GT, Mueller C. Prevalence of delirium and urinary tract infection in a psychogeriatric unit. J Geriatr Psychiatry Neurol 1990;3(4):198–202. [DOI] [PubMed] [Google Scholar]
- 13.Agarwal V, O’Neill PJ, Cotton BA, et al. Prevalence and risk factors for development of delirium in burn intensive care unit patients. J Burn Care Res 2010;31(5):706–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salluh JI, Soares M, Teles JM, et al. Delirium epidemiology in critical care (DECCA): an international study. Crit Care. 2010; 14(6):R210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tuller T, Atar S, Ruppin E, Gurevich M, Achiron A. Common and specific signatures of gene expression and protein-protein interactions in autoimmune diseases. Genes Immun 2013;14(2):67–82. [DOI] [PubMed] [Google Scholar]
- 16.Diao H, Li X, Hu S, Liu Y. Gene expression profiling combined with bioinformatics analysis identify biomarkers for Parkinson disease. PLoS One. 2012;7(12):e52319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park KH, Jung J, Lee JH, Hong YH. Blood transcriptome profiling in myasthenia gravis patients to assess disease activity: a pilot RNA-seq study. Exp Neurobiol 2016;25(1):40–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagalakshmi U, Wang Z, Waern K, et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320(5881):1344–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 2009;10(1):57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirsch J, Vacas S, Terrando N, et al. Perioperative cerebrospinal fluid and plasma inflammatory markers after orthopedic surgery. J Neuroinflammation. 2016;13(1):211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ritter C, Tomasi CD, Dal-Pizzol F, et al. Inflammation biomarkers and delirium in critically ill patients. Crit Care. 2014;18(3): R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Capri M, Yani SL, Chattat R, et al. Preoperative, high-IL-6 blood level is a risk factor of postoperative delirium onset in old patients. Front Endocrinol (Lausanne). 2014;5:173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inouye SK, van Dyck CH, Alessi CA, Balkin S, Siegal AP, Horwitz RI. Clarifying confusion: the confusion assessment method. A new method for detection of delirium. Ann Intern Med 1990; 113(12):941–948. [DOI] [PubMed] [Google Scholar]
- 24.Jorm AF, Scott R, Cullen JS, MacKinnon AJ. Performance of the Informant Questionnaire on Cognitive Decline in the Elderly (IQCODE) as a screening test for dementia. Psychol Med 1991;21(3):785–790. [DOI] [PubMed] [Google Scholar]
- 25.Ruby JG, Bellare P, Derisi JL. PRICE: software for the targeted assembly of components of (meta) genomic sequence data. G3 (Bethesda). 2013;3(5):865–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aken BL, Ayling S, Barrell D, et al. The Ensembl gene annotation system. Database (Oxford). 2016;2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bone RC, Balk RA, Cerra FB, et al. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. The ACCP/SCCM Consensus Conference Committee. American College of Chest Physicians/Society of Critical Care Medicine. Chest 1992;101(6):1644–1655. [DOI] [PubMed] [Google Scholar]
- 30.Benjamini Y, Hochberg Y. Controlling the false discovery rate— a practical and powerful approach to multiple testing. JR Stat Soc Series B Stat Methodol 1995;57(1):289–300. [Google Scholar]
- 31.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 2009;37:W305–W311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ingenuity Upstream Regulator Analysis in IPA. Ingenuity Systems. 2017. http://pages.ingenuity.com/rs/ingenuity/images/0812upstream_regulator_analysis_whitepaper.pdf. Accessed November 2017.
- 33.R: A language and environment for statistical computing [computer program]. Version 3.3.1. Vienna, Austria: R Foundation for Statistical Computing; 2014. [Google Scholar]
- 34.gpots: Various R Programming Tools for Plotting Data. [computer program]. Version: R Package. Vienna, Austria: The R Foundation; 2016. [Google Scholar]
- 35.Platanias LC. Mechanisms of type-I-and type-II-interferon-mediated signalling. Nat Rev Immunol 2005;5(5):375–386. [DOI] [PubMed] [Google Scholar]
- 36.Jorge-Ripper C, Aleman MR, Ros R, et al. Prognostic value of acute delirium recovery in older adults. Geriatr Gerontol Int 2017;17(8):1161–1167. [DOI] [PubMed] [Google Scholar]
- 37.Adamis D, Lunn M, Martin FC, et al. Cytokines and IGF-I in delirious and nondelirious acutely ill older medical inpatients. Age Ageing. 2009;38(3):326–332;discussion 251. [DOI] [PubMed] [Google Scholar]
- 38.Westhoff D, Witlox J, van Aalst C, et al. Preoperative protein profiles in cerebrospinal fluid in elderly hip fracture patients at risk for delirium: a proteomics and validation study. BBA Clin 2015;4:115–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poljak A, Hill M, Hall RJ, et al. Quantitative proteomics of delirium cerebrospinal fluid. Transl Psychiatry. 2014;4:e477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Munster BC, Korevaar JC, Zwinderman AH, Levi M, Wier-singa WJ, De Rooij SE. Time-course of cytokines during delirium in elderly patients with hip fractures. J Am Geriatr Soc 2008; 56(9):1704–1709. [DOI] [PubMed] [Google Scholar]
- 41.van Munster BC, Bisschop PH, Zwinderman AH, et al. Cortisol, interleukins, and S100B in delirium in the elderly. Brain Cogn 2010;74(1):18–23. [DOI] [PubMed] [Google Scholar]
- 42.van Munster BC, Zwinderman AH, de Rooij SE. Genetic variations in the interleukin-6 and interleukin-8 genes and the interleukin-6 receptor gene in delirium. Rejuvenation Res 2011; 14(4):425–428. [DOI] [PubMed] [Google Scholar]
- 43.de Rooij SE, van Munster BC, Korevaar JC, Levi M. Cytokines and acute phase response in delirium. J Psychosom Res 2007; 62(5):521–525. [DOI] [PubMed] [Google Scholar]
- 44.Zaas AK, Chen M, Varkey J, et al. Gene expression signatures diagnose influenza and other symptomatic respiratory viral infections in humans. Cell Host Microbe 2009;6(3):207–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zaas AK, Burke T, Chen M, et al. A host-based RT-PCR gene expression signature to identify acute respiratory viral infection. Sci Transl Med 2013;5(203):203ra126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med 2004;351(27):2817–2826. [DOI] [PubMed] [Google Scholar]
- 47.Klein ME, Dabbs DJ, Shuai Y, et al. Prediction of the Oncotype DX recurrence score: use of pathology-generated equations derived by linear regression analysis. Mod Pathol 2013;26(5):658–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.