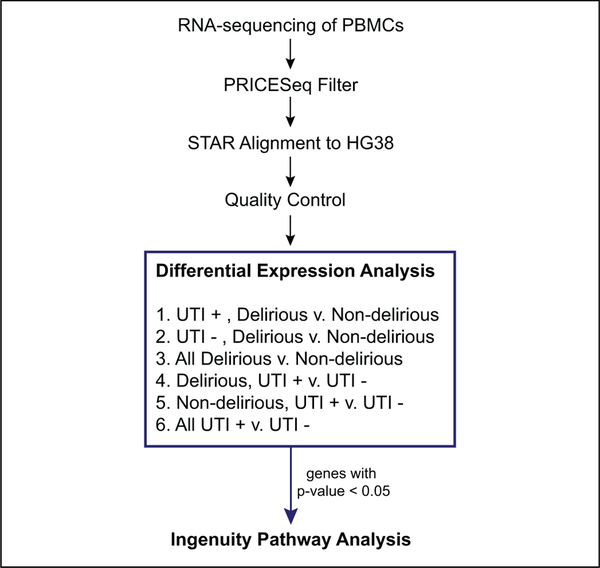
Figure 1.
First, RNA was extracted from PBMCs and sequenced on Illumina HiSeq 2500. The .fastq files obtained from sequencing were quality filtered using PRICESeqFilter and subsequently aligned to HG38 using STAR aligner. After alignment, quality control filters included removal of samples with less than 60% uniquely mapped reads, removal of nonprotein-coding genes, removal of ribosomal and mitochondrial rRNA, and removal of genes with zero read counts in greater than 75% of samples. After quality control, differential expression analysis was performed using DESeq2 on each of the 6 cross sections of the data shown. Genes with raw P value < .05, for each comparison, were included in ingenuity pathways analysis. PBMC indicates peripheral blood mononuclear cell; rRNA, ribosomal ribo-nucleic acid STAR, spliced transcripts alignment to reference.