Figure 2.
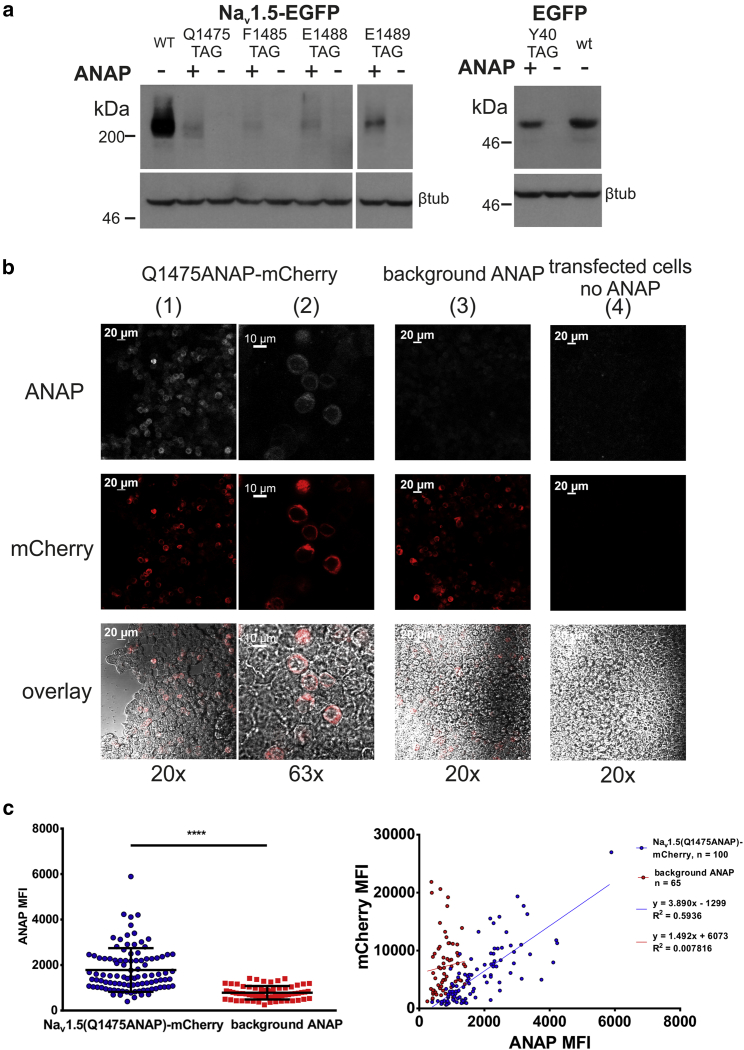
Full-length Nav1.5 expression depends on presence of ANAP and its synthetase-tRNA pair. (a) Western blot of Nav1.5(Q1475TAG)-EGFP and EGFP(Y40TAG) grown in HEK293T ± 10 μM ANAP, probed with rabbit pAb-GFP, is shown. All conditions were co-transfected with AnapRS-tRNALeuCUA. (Left) Nav1.5-EGFP, 5% SDS-PAGE, 5 min exposure is shown. (Right) EGFP, 10% SDS-PAGE, 1 min exposure is shown. Each image in its entirety was adjusted for contrast and brightness enhancement. Full-length EGFP is ∼27 kDa and was observed ∼50 kDa in this blot. Full-length Nav1.5-EGFP is ∼275 kDa and was observed above 200 kDa in this blot. (b) Fluorescence imaging of HEK293T expressing Nav1.5(Q1475ANAP)-mCherry (columns 1, 2), Nav1.5-mCherry+10 μM ANAP (background ANAP, column 3), or Nav1.5(Q1475TAG)-mCherry in the absence of ANAP (transfected cells, no ANAP, column 4) is shown. ANAP (ex405, em445/50 nm) and mCherry (ex555, em647/70 nm) fluorescence were imaged. Overlay image includes ANAP (white), mCherry (red), and brightfield channels. All except brightfield images were equally adjusted in their entirety for contrast enhancement. Columns 1, 3, and 4 scale bars represent 20 μm. Column 2 scale bars represent 10 μm. (c) Quantification of the 20× images in (b) is shown. (Left) ANAP mean fluorescence intensity (MFI) was significantly increased when the target codon for incorporation was present. Error bars are ± standard deviation. ∗∗∗∗p < 0.0001, two-tailed unpaired t-test with Welch’s correction. (Right) mCherry MFI plotted as a function of ANAP MFI is shown. To see this figure in color, go online.