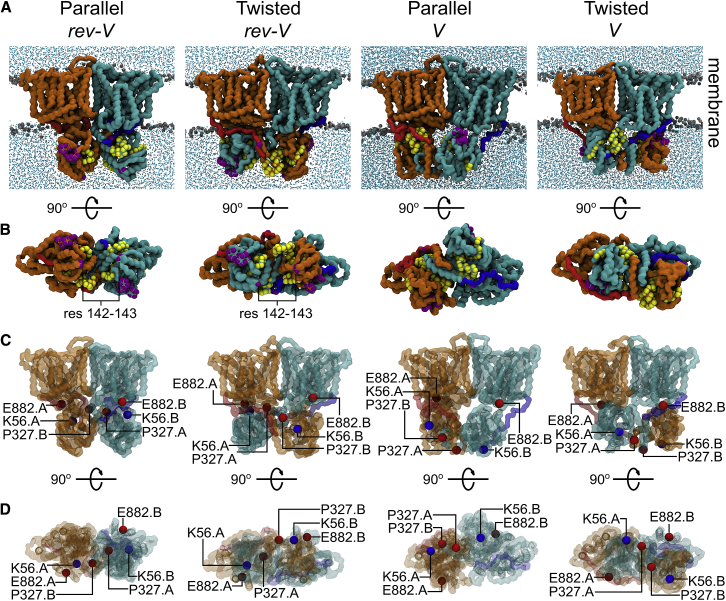
Figure 2.
The AE1 configurations resulting from the simulations. (A) Structures represent the most sampled conformation (i.e., a centroid) for each conformer in the CG-MD simulations. The surface of the backbone particles of the AE1 complex dimer subunits are shown in orange and cyan with the linker regions in red and blue. Phosphate groups from the lipid bilayer and water molecules are shown in gray and cyan, respectively. (B) Shown is the cytoplasmic view of the systems in (A). Residues shown in other studies to interact with actin (residues 258–311) and ankyrin (63–73, 142, 143, 175–185, 353) are shown as yellow and purple spheres, respectively. (C) The position of the variants Memphis (K56E) (18, 64, 65) and Tuscaloosa (P327R) (20) is shown on the structures from A as blue and red spheres, respectively. (D) Shown are the cytoplasmic views of the systems in (C). The E882 residue, closer to the Tuscaloosa variant in the rev-V conformers, is also shown. See also Figs. S1 and S3–S8. To see this figure in color, go online.