Figure 3.
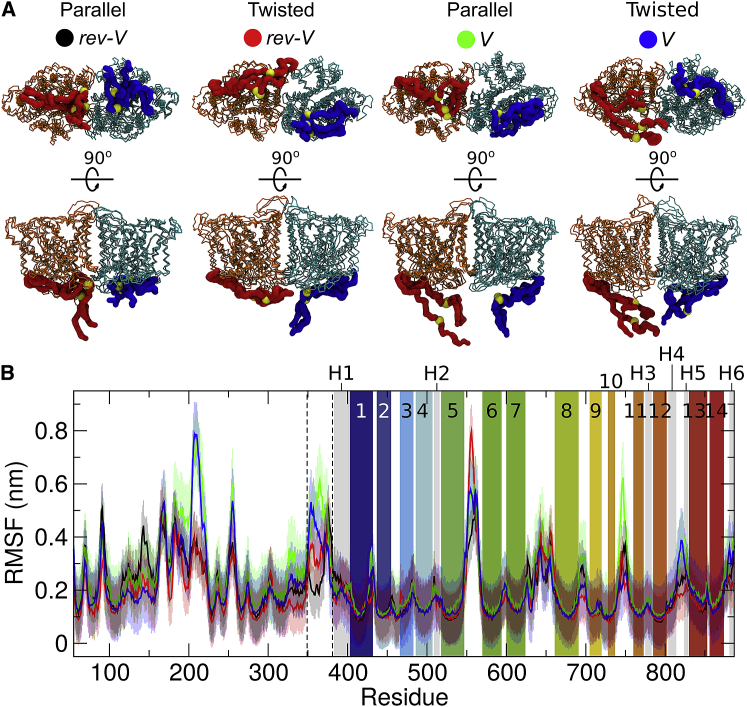
AE1 linker position. (A) The AE1 structures are the three most populated centroids after the cluster analyses of the CG-MD simulations. The protein has been aligned on the mdAE1 backbone atoms. The backbone particles of the mdAE1 are shown in orange and cyan with the linker regions in red and blue. The residue K360 is shown as a yellow sphere. The cdAE1 domain has been omitted for clarity. (B) Shown is the root mean-square fluctuation (RMSF) of the intact AE1 in the atomistic simulations. The mdAE1 TM helices (TM1 to TM14) are highlighted in rainbow, and the reentrant helices are in gray (H1 to H6). The residues of the linker regions are shown between the dotted lines. Each line represents the average and the SD over the three repeat atomistic simulations of each system. To see this figure in color, go online.