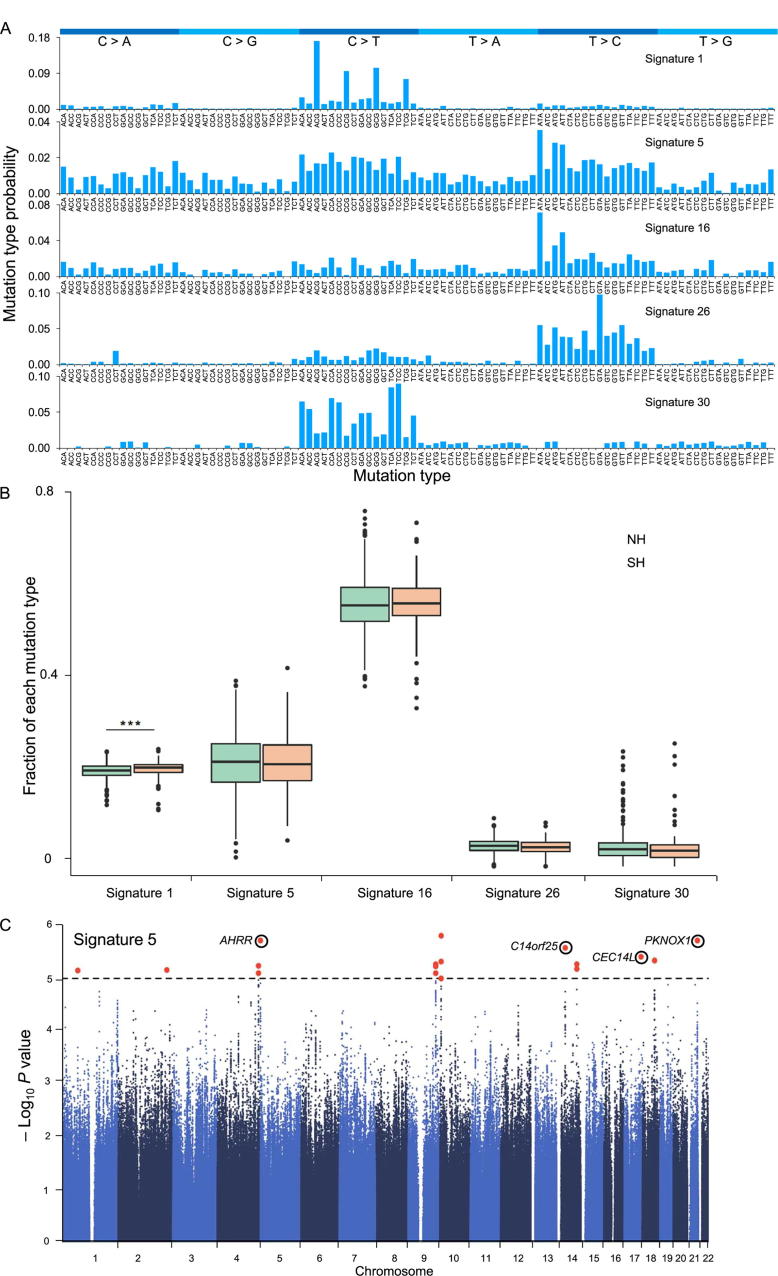
Figure 4.
The population distribution of mutational signatures
A. Five COSMIC mutation signatures with patterns matching analysis of the novel singletons identified in the CASPMI cohort. The 96 types of trinucleotide mutational contexts are presented on the x axis, and y-axis shows the probability of a specific mutation occurring in such a context. B. Distribution of the five aforementioned mutational signatures in the NH and SH groups. Signature 1 showed the most significant difference between these 2 groups (P = 0.001, Wilcox rank test). Boxplots show the proportion of each mutational signature in NH (green) and SH (orange) individuals. Whiskers denote the lowest and highest values within 1.5 times the range of the first and third quartiles, respectively; dots represent outliers beyond the whiskers. C. SNPs significantly associated with the individual load of COSMIC signature 5. 17 significant SNPs were identified as being associated with the individual load of this signature (P < 10−5). Dashed horizontal line represents the significance threshold (P = 10−5). Red dots represent the significant SNPs, and black circles indicate the genes where the significant SNPs reside. COSMIC, the Catalogue of Somatic Mutations in Cancer.