Figure 5.
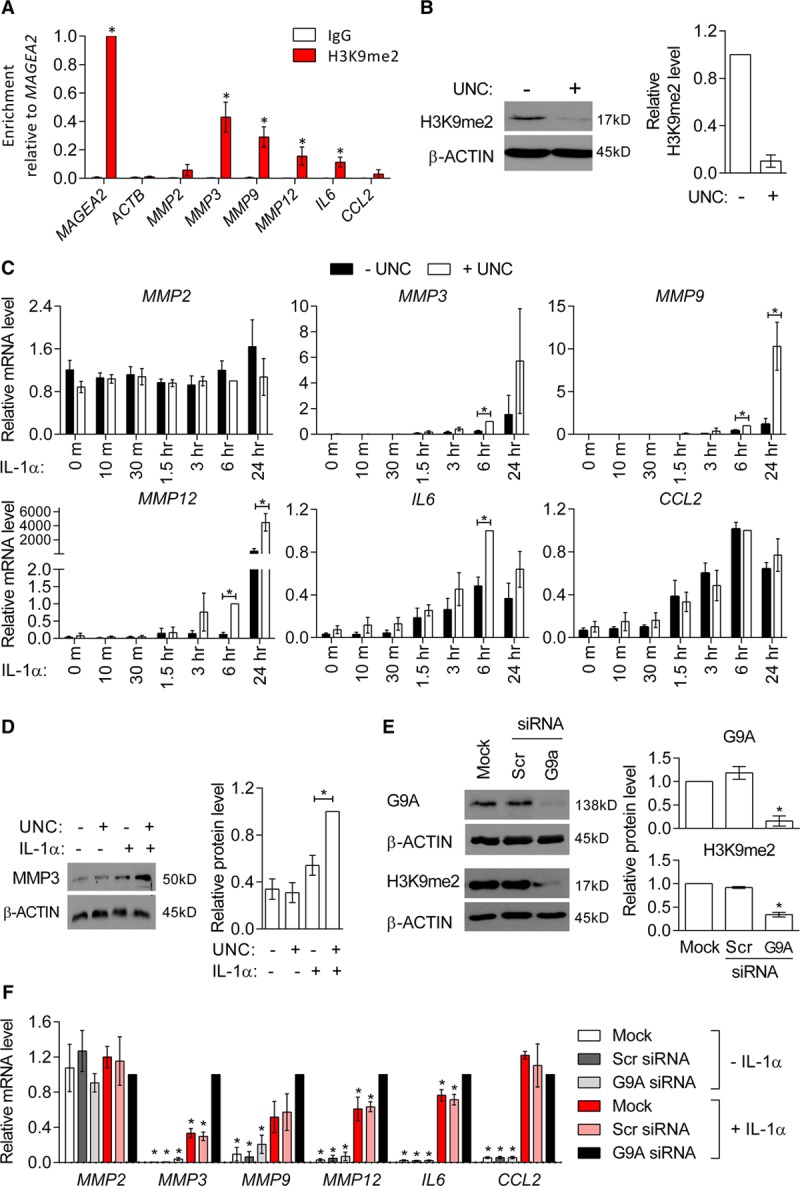
Histone H3 lysine 9 dimethylation (H3K9me2)-mediated regulation of inflammation-associated genes is conserved in human aortic VSMCs (hVSMCs). A, Chromatin immunoprecipitation-qPCR analysis in cultured hVSMCs, showing enrichment for H3K9me2 relative to MAGEA2 (positive control) at ACTB (negative control) and MMP2, MMP3, MMP9, IL6, and CCL2 promoters compared with signals for negative control IgG (immunoglobulin). Graph show mean±SEM of 6 experiments. *P<0.05 (Kruskal-Wallis, H3K9me2 enrichment compared with ACTB). B, Representative western blot and densitometric analysis of H3K9me2 levels in control (−UNC [UNC0638]) and UNC-treated (+UNC) cultured hVSMCs. Data (mean±SEM of 3 experiments) are shown relative to untreated cells and normalized to β-actin. C, RT-qPCR (reverse transcription with quantitative polymerase chain reaction) analysis of MMP2, MMP3, MMP9, MMP12, IL6, and CCL2 in control hVSMCs and after IL-1α treatment for 10, 30 min, 1.5, 3, 6, or 24 h, without (−UNC, black bars) and with prior UNC treatment (+UNC, white bars). Data are relative to cells treated with UNC+IL-1α for 6 h and normalized to housekeeping genes (HMBS and YWHAZ). Graph show mean±SEM of 4 to 10 experiments. *P<0.05 (Kruskal-Wallis). D, Representative western blot and densitomeric analysis of MMP3 in control, UNC, IL-1α, and UNC+IL-1α-treated cultured hVSMCs. Mean±SEM of 3 experiments is shown relative to cells treated with UNC+IL-1α and normalized to β-actin. *P<0.05 (Kruskal-Wallis). E, Representative western blot and densitometric analysis of G9A and H3K9me2 levels in mock, scrambled (Scr) siRNA, and G9A siRNA transfected hVSMCs. Data are shown relative to mock-transfected hVSMCs and normalized to β-actin (mean±SEM of 4 experiments). *P<0.05 (Kruskal-Wallis, compared with Scr). F, RT-qPCR analysis of MMP2, MMP3, MMP9, MMP12, IL6, and CCL2 in mock, Scr siRNA and G9A siRNA-transfected hVSMCs with and without IL-1α stimulation. Data (mean±SEM of 4 experiments) are relative to hVSMCs treated with G9A siRNA+IL-1α and normalized to housekeeping genes (HPRT1 and YWHAZ). *P<0.05 (Kruskal-Wallis, compared with G9A siRNA+IL-1α). Each experiment used hVSMC isolates derived from different individuals.
