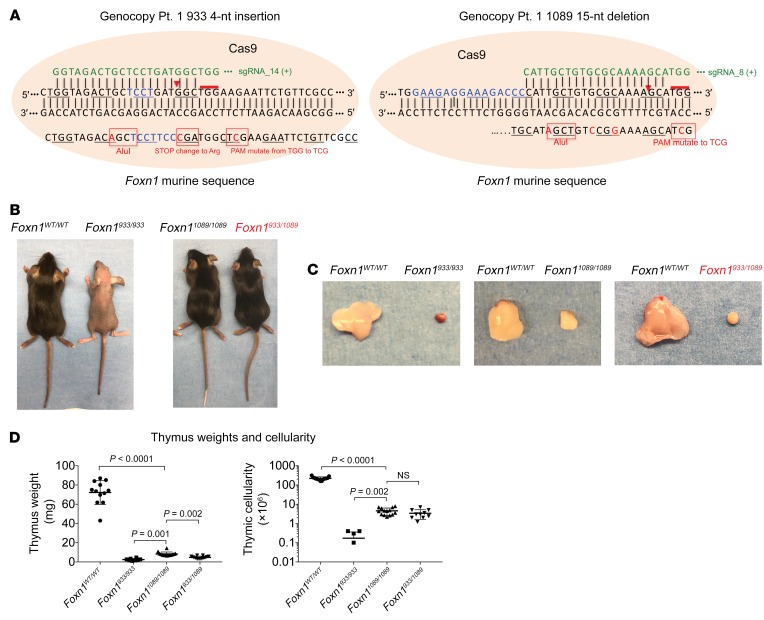
Figure 2. Human FOXN1 compound heterozygous mutations genocopied in mice cause thymic aplasia with normal fur and nails.
(A) The human FOXN1 mutations for Pt. 1 were introduced into the murine genome by CRISPR-Cas9 technology. The DNA repair template used for each allele is shown. Silent mutations were introduced into the murine sequence to facilitate genotyping and to prevent premature stop codons. (B) Images of F2-generation mice: WT Foxn1 (Foxn1WT/WT), homozygous mutant (Foxn1933/933 and Foxn11089/1089), and compound heterozygous (Foxn1933/1089) mice, the latter genocopying Pt. 1. The genocopied mice are indicated in red font. The images are representative of 5 independently characterized mice. (C) The overall sizes of the thymi from the various mouse lines are shown for comparative purposes. (D) Thymus weights and overall thymic cellularity were calculated. Data represent the mean ± SEM. n = 22 Foxn1WT/WT, n = 6 Foxn1933/933, n = 10 Foxn11089/1089, and n = 10 Foxn1933/1089 mice. P values of less than 0.05 were considered significant. For the comparisons shown, a Brown-Forsythe and Welch’s 1-way ANOVA was applied.