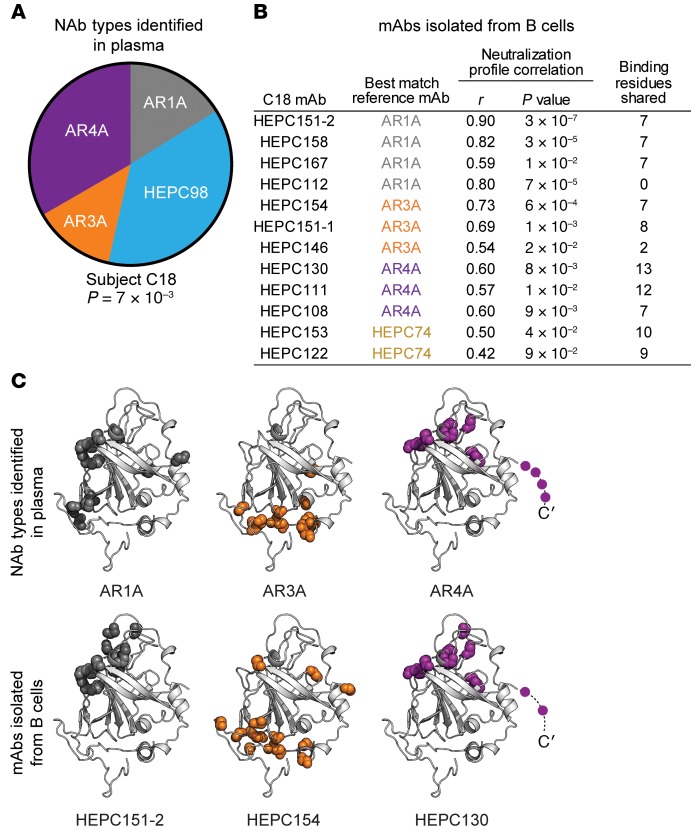
Figure 5. Plasma NAb deconvolution predicts mAbs subsequently isolated from B cells of the same subject.
(A) NAb deconvolution of subject C18 plasma. Plasma neutralization profile was averaged from 2 independent experiments. Reference mAb profiles were averaged from 5 independent experiments. Wedge sizes are proportional to the plasma response attributed to each reference mAb. P value is from the Pearson correlation of the C18 plasma neutralization profile with a combined neutralization profile comprising the indicated proportion of each reference mAb. (B) Pearson correlations between neutralization profiles and number of shared probable E1/E2-binding residues, as determined by alanine-scanning mutagenesis–E1/E2-binding analysis, between 12 mAbs isolated from subject C18 B cells and best-match reference mAbs. C18 mAb neutralization profiles were determined in a single experiment, with neutralization of each HCVpp measured in duplicate. (C) Examples of concordance between binding residues of reference mAbs identified in plasma and 3 best-match mAbs isolated from C18 B cells. Colored spheres indicated main chain atoms of probable binding residues, superimposed on the crystallized structure of the HCV E2 protein ectodomain, strain 1b09, PDB accession 6MEI (15), with colors modified in PyMOL, v1.8.6.2. Shared putative E1 binding residues of HEPC130 and AR4A (Y201, N205) are not shown.