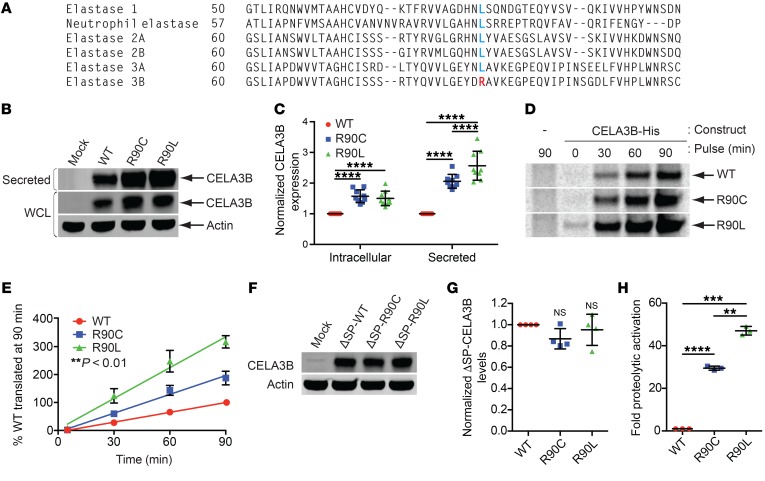
Figure 2. Functional characterization of CELA3B mutations at arginine 90.
(A) Sequence alignment of the 6 human elastases centered at position 90 in CELA3B. Arginine (R) residues are labeled in red; leucine (L) residues are labeled in blue. (B) Immunoblots of whole-cell lysates (intracellular) and conditioned media (secreted protein) from 293T cells transfected with empty vector (Mock) or the indicated CELA3B variant. (C) Quantification from the 9 experiments in B. Values represent the mean ± SD. (D) Autoradiographs of His-tagged CELA3B variants purified from brefeldin A–treated 293T cells pulsed with 35S-labeled methionine and cysteine for the indicated durations. The first (control) lane is the same in the WT and R90C images. (E) Quantification from the 4 experiments in D, showing data points normalized to WT levels at 90 minutes and their corresponding lines of best fit (linear regression). Values represent the mean ± SD. **P < 0.01 among all samples. (F) Western blots of CELA3B variants lacking a signal peptide (ΔSP). Samples are whole-cell lysates from 293T cells transfected with empty vector (Mock) or vector containing ΔSP-CELA3B variants. Blots were probed with the indicated antibodies. (G) Quantification of data from the 4 experiments in F, with protein levels normalized to ΔSP-WT CELA3B. Values represent the mean ± SD. (H) Efficiency of catalytic activation of CELA3B variants in conditioned media after limited activation with trypsin. Raw catalytic activity was quantified as a change in absorbance due to cleavage of a colorimetric CELA3B substrate. Raw values were normalized to total CELA3B protein levels and are shown as the mean ± SD for 3 experiments. Multiplicity-adjusted P values were determined by matched 2-way ANOVA (C) or matched 1-way ANOVA (panels G and H) with Tukey’s multiple comparisons test. **P < 0.01, ***P < 0.001, and ****P < 0.0001.