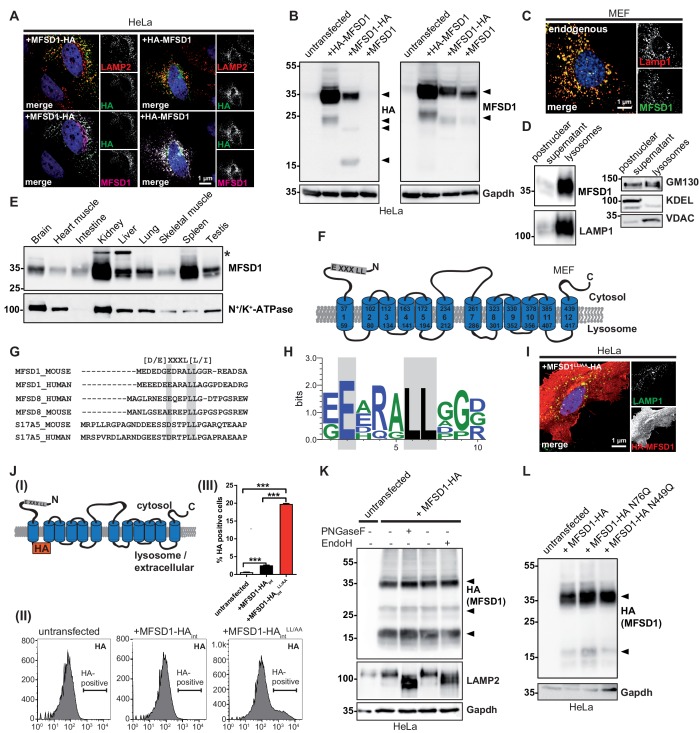
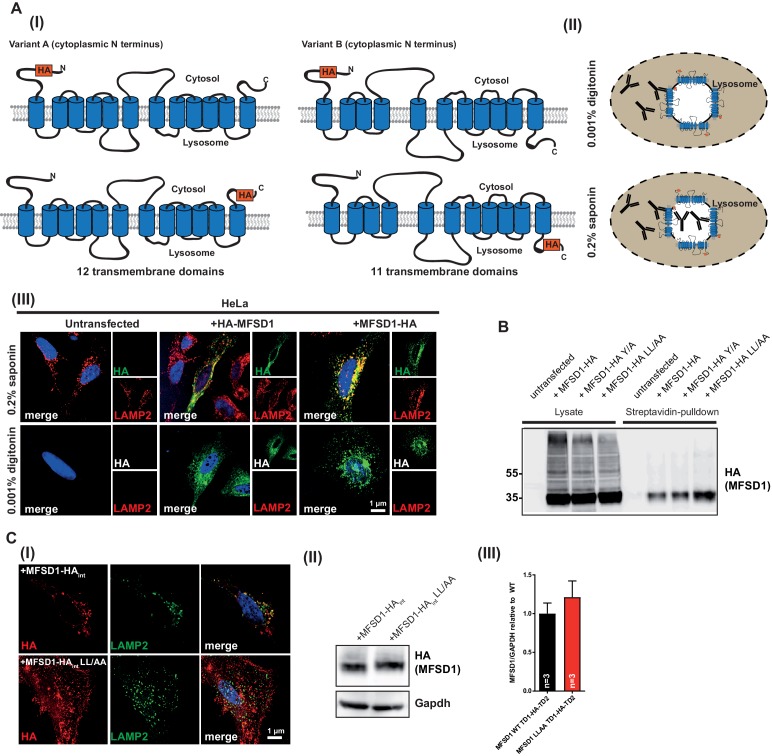
Figure 1. MFSD1 is an ubiquitously expressed, non-glycosylated lysosomal protein that contains a dileucine-based lysosomal sorting motif.
(A) Co-Immunofluorescence staining of HeLa cells overexpressing N- or C-terminally HA-tagged MFSD1 with antibodies against LAMP2 (red), HA (green) and an antibody against MFSD1 (magenta). (B) Immunoblot of HeLa cells transfected with untagged and N- or C-terminally tagged MFSD1 with antibodies against HA (left panel) and MFSD1 (right panel). Gapdh is depicted as loading control. MFSD1-specific bands are labeled with arrow-heads. (C) Co-immunofluorescence staining for LAMP1 (red) and endogenous MFSD1 (green) of wildtype MEF cells. (D) Immunoblot of the postnuclear supernatant and the lysosome-enriched fraction of magnetite-isolated lysosomes from wildtype MEFs probed with antibodies against MFSD1, LAMP1 (marker for lysosomes), GM130 (marker for the Golgi-apparatus), KDEL (marker for the endoplasmic reticulum) and VDAC (marker for mitochondria). (E) Immunoblot of crude membranes of the indicated wildtype mouse tissues probed with the antibody against MFSD1. N+/K+-ATPase is depicted as a loading control. An unspecific band is labeled with an asterisk. (F) Topology model of murine MFSD1. The putative dileucine-based lysosomal sorting motif is indicated. Amino acids confining the TMDs (numbered from 1 to 12) are indicated. (G) Alignment of the dileucine-based lysosomal sorting motifs-containing N-termini of the indicated proteins/species. The critical amino acids constituting the motif are depicted in gray. (H) Web-logo representation of the sequence containing the dileucine motif of MFSD1 orthologues from different species (Mus musculus, Rattus norvegicus, Homo sapiens, Bos taurus, Gallus gallus, Danio Rerio). (I) Co-immunofluorescence for LAMP2 (green) and HA (red) of HeLa cells transfected with N-terminally HA-tagged MFSD1LL/AA-mutant. (J) (I) Schematic representation of the MFSD1-construct with an internal-HA-tag between TMD 1 and 2 used for FACS. (II) FACS plots of untransfected HeLa cells, cells transfected with wildtype and MFSD1LL/AA-mutant with an internal HA tag after staining of HA without permeabilization. (III) Quantification of HA-positive cells from three FACS experiments. ***=p < 0.001. (K) Immunoblot of lysates from MFSD1-HA-transfected HeLa cells treated with PNGaseF or EndoH probed with an antibody against HA. Gapdh and LAMP2 are shown as loading controls and controls for efficient glycosidase-treatment, respectively. (L) Immunoblot of HeLa cells transfected with wildtype C-terminally HA-tagged MFSD1 or the indicated mutants N76Q/N449Q probed with an antibody against HA. Gapdh is shown as loading control.