Abstract
We provide protocols for titering and isolating bacterial colonies from single cells by serial dilutions, for streaking agar plates, and for spreading suspensions of cells on plates. Support protocols describe replica plating, and methods for storing strains as agar stabs and frozen stocks.
BASIC PROTOCOL 1
TITERING AND ISOLATING BACTERIAL COLONIES BY SERIAL DILUTIONS
Bacteria are grown from single colonies to ensure that each cell in a population is descended from a single founder cell. Cultures grown from single cells are clonal, that is, with the exception of mutations that occurred since division of the founder cell, each cell in the culture has the same genetic makeup. One way to generate single colonies is to titer a culture with serial dilutions and to pick colonies from one of the dilution plates. In this procedure, a small, measured amount of a bacterial culture is diluted into fresh liquid in another tube. A small amount of liquid is taken from this tube and diluted into another fresh tube. This process is repeated several times. Equal volumes of liquid are then taken from each of the dilution tubes and plated on petri plates. The plates are incubated overnight at 37°C; well-separated single colonies will arise on some of the dilution plates.
Isolation of single colonies by this means also allows calculation of the number of cells present in the starting culture from the number of colonies formed on the dilution plates. The process of determining the concentration of entities in liquid by serial dilution followed by detection is called titration.
A typical saturated culture of E. coli. contains 1 × 109 cells/ml. Phage suspensions can also be titered; a concentrated stock of a λ derived phage might typically contain 1 × 1011 phage/ml.
Materials
LB medium (unit 1.1)
LB plates (unit 1.1)
Sterile 16- or 18-mm-diameter culture tubes
Use pipets to introduce 5 ml LB medium into three sterile culture tubes. Line the tubes up, or label them so that they can be distinguished.
Using a pipettor, transfer 5 μl from the suspension of cells into the first tube of LB medium. Set the vortexer to a mild setting and agitate the tube 5 sec.
Put a new tip on the pipettor and transfer 5 μl from the first tube of LB medium into the second tube, and vortex the second tube. Take 5 μl from the second tube and repeat step 3 until you have serial dilutions in all three tubes.
The first dilution tube now contains a 1 × 103-fold dilution, generated by diluting the culture by a factor of one thousand (i.e., it contains 1 × 10−3 as many cells/ml as were present in the original culture). The second tube contains a 1 × 106-fold dilution, generated by diluting the original culture by a factor of one million (i.e., it contains 1 × 10−6 as many cells/ml as the original culture), etc.
Many investigators prefer to perform serial dilutions with different volumes and different factors of dilution. These parameters can be modified in steps 1 to 3.
Spread 100 μl of liquid from the culture and from each dilution tube onto separate, labeled, dry LB plates (as described on p. 1.3.2). Incubate overnight at 37°C.
During this incubation, each living bacterial cell will grow into a separate colony on the plate.
Count the colonies from these plates. Since only 100 μl was plated from the undiluted culture and from each dilution tube, each plate has 1/10 as many colonies on it as were present in each milliliter of liquid in the corresponding tube. Therefore, one can determine the number of cells that were present per milliliter of the culture by counting the number of colonies on a plate, and then multiplying that number by 10 times the factor of dilution.
For example, if 22 colonies were observed on the plate corresponding to the 1 × 106-fold dilution, then the number of living cells in each milliliter of the original culture was 22 × 10 ×106, or 2.2 ×108 cells/ml.
Any of the single colonies may be saved for further use. Store plates at 4°C wrapped in parafilm or in the plastic sleeve in which the plates were supplied.
Commentary
Titering by serial dilutions is a good way to determine the number of any kind of living organism present in a suspension. The organisms do not even need to be able to grow into colonies—i.e., the concentration of living bacteriophage in a tube can be determined by titering with serial dilutions and counting the number of plaques made when an aliquot of each dilution is plated on a lawn of phage-sensitive bacteria (see unit 1.11).
It is sometimes useful to use smaller factors of dilution. Mixing 50 μl of the culture into 5 ml medium will give dilutions of 100×. Mixing 100 μl into 900 μl will give dilutions of 10×. Mixing 315μl into 685 μl will give dilutions of 3.15×, which are sometimes useful because two such steps result in dilution by a factor of 10.
BASIC PROTOCOL 2
ISOLATING SINGLE COLONIES BY STREAKING A PLATE
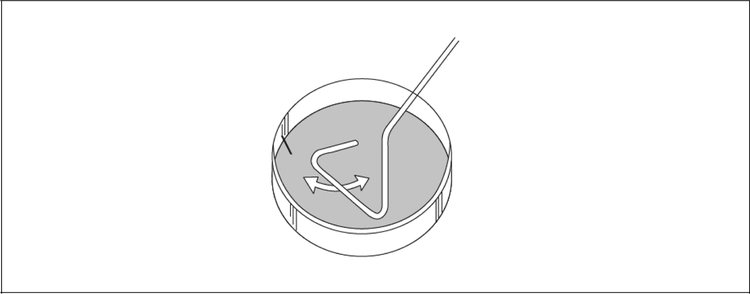
Another way to isolate single colonies is called streaking or streaking for single colonies. For isolatiing single colonies, this method is easier and faster than serial dilutions for isolating single colonies, but it cannot be used to count the number of cells in a culture. An inoculum of bacteria is streaked across one side of an agar plate with an inoculating loop or sterile toothpick. The resterilized loop or a fresh toothpick is then passed once through the first streak and streaked across a fresh part of the plate (see Fig. 1). This process is repeated at least once more, and the plate is incubated at 37°C until colonies become visible. If single colonies must be isolated from many bacteria, it is convenient to divide a plate into 4, 6, or 8 sectors and to streak for single colonies in each sector.
Figure 1.
A plate streaked for single colonies. Sinusoidal marks show path traced by successive streaks of a dowel, toothpick, or inoculating loop.
BASIC PROTOCOL 3
ISOLATING SINGLE COLONIES BY SPREADING A PLATE
It is sometimes necessary to distribute a liquid culture of bacteria evenly over the surface of a plate (for example, when plasmid-containing colonies are to be isolated after transformation of cells with plasmid DNA and calcium chloride, unit 1.8). This is usually done with a glass spreader. From 0.05 to 1 ml of liquid is pipetted onto a dry plate (see Elbing And Brent, UNIT 1) and spread using a circular motion as shown in Figure 2. Alternatively, the edge of the spreader can be used to make a raster pattern on the plate’s surface. The plate can be turned at right angles and the process repeated. In another alternative approach, the spreader can be moved back and forth across the plate while the plate is spun slowly on a turntable. Evenly spread plates should be placed in the incubator with the lids ajar until they are completely dry.
Figure 2.
Spreading a plate.
SUPPORT PROTOCOL 1
REPLICA PLATING
Replica plating is a convenient way to test many colonies for their ability to grow under different conditions. In this technique, bacterial colonies are transferred from one plate to another in a way that maintains the original pattern of colonies. This technique has many applications to recombinant DNA work. As an example, consider the plasmid pBR322, which contains two antibiotic resistance genes, encoding resistance to ampicillin and tetracycline (see Figure in Unit 1.5). A piece of foreign DNA cloned into the tetracycline resistance gene inactivates it; cells that carry such a plasmid are ampicillin resistant but tetracycline sensitive. These cells can be identified by replica plating colonies from ampicillin-containing master plates onto plates containing tetracyline. Tetracyline-sensitive colonies can be identified by their inability to grow on the tetracycline plates, rescued from the master plate, and analyzed further. If desired, an ampicillin-containing plate can also be included as a positive control to demonstrate adequate cell transfer.
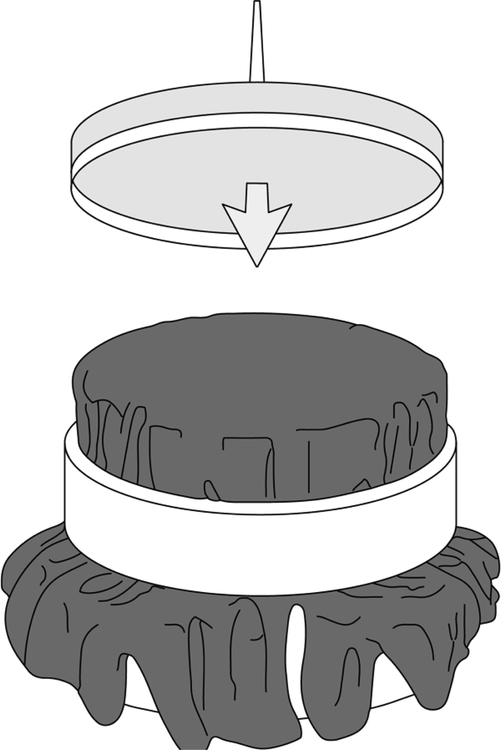
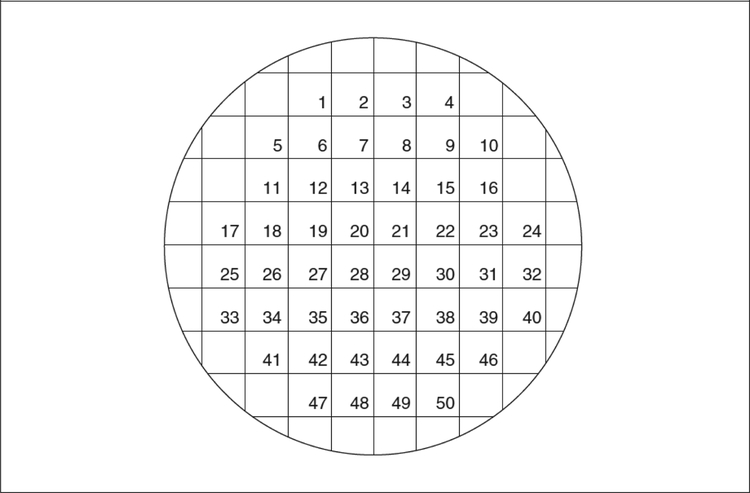
This procedure requires two specialty items: a replica block and sterile velvets. The replica block is a wooden or metal cylinder that fits snugly inside a petri plate (see Fig. 3). One method for constructing these from mid-20th century automobile pistons has been described by Adams (1965). A metal ring is used to secure the velvets to the block. Squares of velvet should be cut so as to cover the base (a diameter of 14 cm is suggested). These velvets can be washed, autoclaved, and reused. If velvets are not available, pieces of sterile filter paper or disposable replica plates can be used (“Repli-Plate” Colony Transfer Pads, American Laboratories #59901). Replica plating also requires a master plate composed of well-separated colonies. The master plate can be a fresh plate onto which 50 to 100 colonies have been gridded (using toothpicks and the grid in Fig. 4), or it can be a plate on which bacteria were spread that have now grown up into well-separated colonies.
Figure 3.
Replica block with velvet. Petri dish with agar is being pressed down onto the velvet.
Figure 4.
Grid for replica plating.
Mark the top of the master plate to enable alignment with the grid. Press the plate down lightly onto the velvet. Do not bear down hard on the plate; pressing too hard will cause the colonies to run together on the velvet or may even cause the plate to collapse. Press new plates, oriented like the master plate, lightly onto the imprinted velvet to transfer the colonies. As many as 10 plates per velvet can often be replica plated.
SUPPORT PROTOCOL 2
STRAIN STORAGE AND REVIVAL
Most strains of E. coli can be stored for years in stab vials, or indefinitely if frozen at −70–80°C. It is prudent to check the genetic markers of a strain revived from storage. Ways to verify the presence of many different selective markers are described in Unit 1.4, Table 1.4.4.
Stabs
Use airtight, autoclavable vials with rubber or Teflon caps (not cardboard). These are available from Wheaton Glassware and John’s Scientific (¼-oz. Bijoux bottles, #15690001). Fill the vials ⅔ full with stab agar (unit 1.1). Inoculate them with a single colony (see Fig. 1.3.5) as follows. Collect most of the cells in the colony with an inoculating loop, then repeatedly poke the loop deeply into the agar. Replace the screw cap of the stab vial. Leave the cap of the stab vial slightly loose and incubate 8 to 12 hr at 37°C, or until cloudy tracks of bacterial growth are evident. Seal the vials tightly and store them in a cool (15° to 22°C), dark place. To revive a stored strain, flame sterilize an inoculating loop (Elbing and Brent companion paper, unit 1.1), allow it to cool, insert it into the stab agar, and move the loop around until a gobbet of bacteria-laden agar is stuck onto the loop. Smear the gobbet onto one section of an LB plate and streak for single colonies (Fig. 1).
Frozen Stocks
Add 2 ml of a mid-log culture or 1 ml of a freshly saturated culture to a stab vial or a Nunc vial (Nunc #1087) containing 1 ml glycerol solution or DMSO solution (see recipes). Vials can be stored at −20° to −70°C, but most strains remain viable longer if stored at −70°C. Revive stored cells by scraping off splinters of solid ice with a steriel toothpick, applicator stick, or pipet and streaking these splinters onto an LB plate (Elbing and Brent, unit 1.1). Do not allow the contents of the vial to thaw.
REAGENTS AND SOLUTIONS
Use deionized, distilled water in all recipes and protocol steps. For common stock solutions, see Current Protocols in Molecular Biology, appendix 2; for suppliers, see Current Protocols in Molecular Biology, appendix 4.
Glycerol solution
65% glycerol (vol/vol)
0.1 M MgSO4
0.025 M Tris Cl, pH 8
DMSO solution
7% dimethylsulfoxide (vol/vol)
The only advantage DMSO seems to have over glycerol for frozen stocks is that it is easier to pipet because it is less viscous. Use a bottle of reagent- or spectrophotometric-grade DMSO that has been kept tightly sealed. Old bottles of DMSO sometimes develops a mercaptan or sulfurish odor. If the DMSO has any smell, do not use it.
Figure 5.
Using wire loop to inoculate a stab vial. Tracks in the agar show previous stabs with the loop.
Acknowledgement
This work was supported by NIH grant R21 CA223901 to RB.
Contributor Information
Karen Elbing, Clark & Elbing LLP, Boston, Massachusetts.
Roger Brent, Division of Basic Sciences, Fred Hutchinston Cancer Research Center, Seattle, Washington, rbrent@fredhutch.org.
REFERENCES
- Adams JN 1965. Automotive pistons for use as bases in velveteen replication. J. Bacteriol 89: 1627. [DOI] [PMC free article] [PubMed] [Google Scholar]