Figure 6. The YAP-β-catenin signaling targets XBP1 and inhibits NLRP3-driven inflammatory response in MSC-mediated immune regulation.
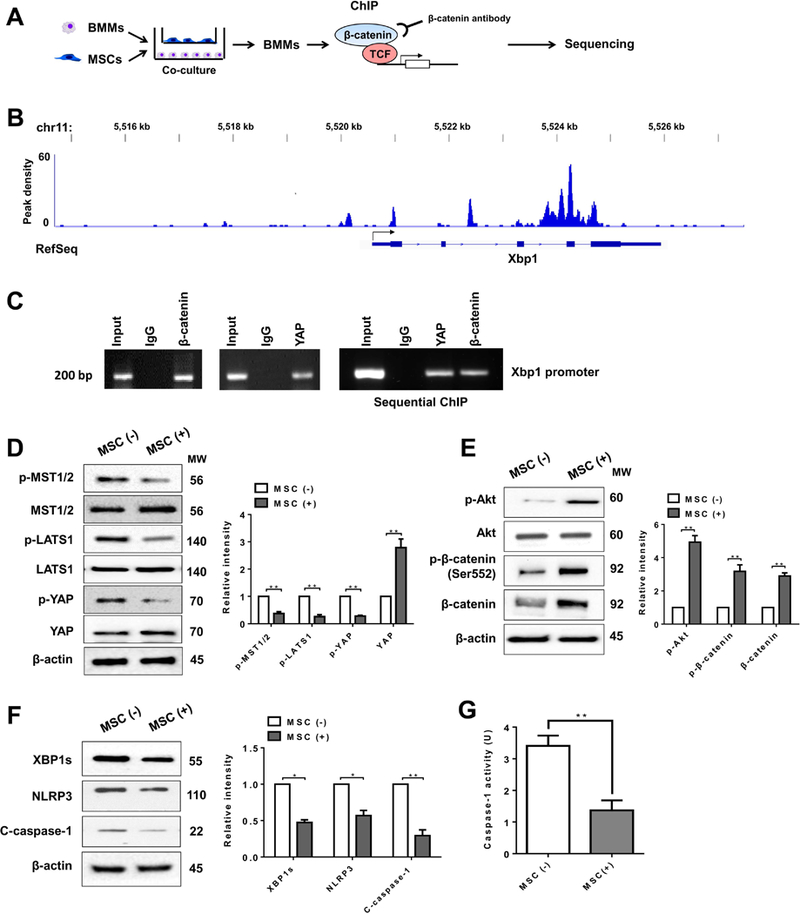
(A) Experimental design of β-catenin ChIP-seq analysis. BMMs were collected and fixed after co-culture with MSCs. Following chromatin shearing and β-catenin antibody selection, the precipitated DNA fragments bound by β-catenin-containing protein complexes were used for sequencing. (B) Localization of β-catenin-binding sites on the mouse xbp1 gene. The five exons, four introns, 3’ UTR, 5’ UTR and TSS of the mouse xbp1 gene on chromosome 11 are shown. (C) ChIP-PCR analysis of YAP and β-catenin binding to the Xbp1 promoter. Protein-bound chromatin was prepared from BMMs and immunoprecipitated with YAP or β-catenin antibodies. For sequential ChIP, the protein-bound chromatin was first immunoprecipitated with the β-catenin antibody followed by elution with a second immunoprecipitation using YAP antibody, and then the immunoprecipitated DNA was analyzed by PCR. The normal IgG was used as a negative control. (D) (E) (F) Immunoblot-assisted analysis and relative density ratio of p-MST½, MST½, p-LATS1, LATS1, p-YAP, YAP, p-Akt, Akt, p-β-catenin, β-catenin, XBP1s, NLRP3, and cleaved caspase-1 in macrophages after co-culture with or without MSCs. Representative of three experiments. (G) Caspase-1 activity (U) in macrophages after co-culture (n=3–4 samples/group). All data represent the mean±SD. *p<0.05, **p<0.01.
